Fungal NAD+ metabolism gets extracellular
Published in Microbiology

We live in a time where viral and bacterial infectious diseases have become conversation topics as normal as the weather, but diseases caused by fungi still receive little attention even in the scientific community. 300 million people suffer from fungal-related disorders each year, resulting in more than 1.6 million deaths; a number that is greater than those attributed to malaria (Nature Microbiology 2:17120). Among the fungi able to cause diseases in humans, representatives of the species Aspergillus, Candida, Cryptococcus, and Pneumocystis are the most common causes of deadly fungal infections. At the Leibniz Institute for Natural Product Research and Infection Biology (HKI), Department of Molecular and Applied Microbiology (MAM), we dedicate our research to studying the infection biology of the human-pathogenic mould Aspergillus fumigatus. This thermotolerant fungus can be found worldwide in the soil, where it degrades organic matter. It propagates via spores (termed conidia), which can be easily inhaled by humans due to their small size of only 2-3 µm. Healthy individuals are usually not affected, but patients with severely impaired immune systems, e.g. after a bone marrow transplantation, are at risk of suffering a life-threatening, invasive infection, invasive aspergillosis. In addition, A. fumigatus also causes allergic diseases (Latgé and Chamillos 2019).
In our lab, we aim to obtain a better understanding of the pathophysiology of A. fumigatus and work towards developing new diagnostic and therapeutic options. In particular, proteins present on the conidial surface play an important role during host-pathogen interaction by mediating adhesion or immune evasion (Blango et al. 2019). A few years ago, our lab discovered the surface protein CcpA by proteome analyses. CcpA is highly abundant on dormant conidia of A. fumigatus and further experiments demonstrated its contribution to virulence, since it has a structural function and masks conidial antigens from recognition by the innate immune system (Voltersen et al. 2018). Encouraged by these results, we further characterized the conidial surface proteins of A. fumigatus and other moulds. Our preferred approach is a short treatment of intact conidia with trypsin to release surface exposed peptides without damaging the conidia. This method, known as “surface shaving”, has been successfully used to identify surface-associated proteins of all kinds of microorganisms, including fungi. Many of the detected surface proteins are hardly characterized and are not assigned to any functional group. These experiments highlight how much we still have to discover!
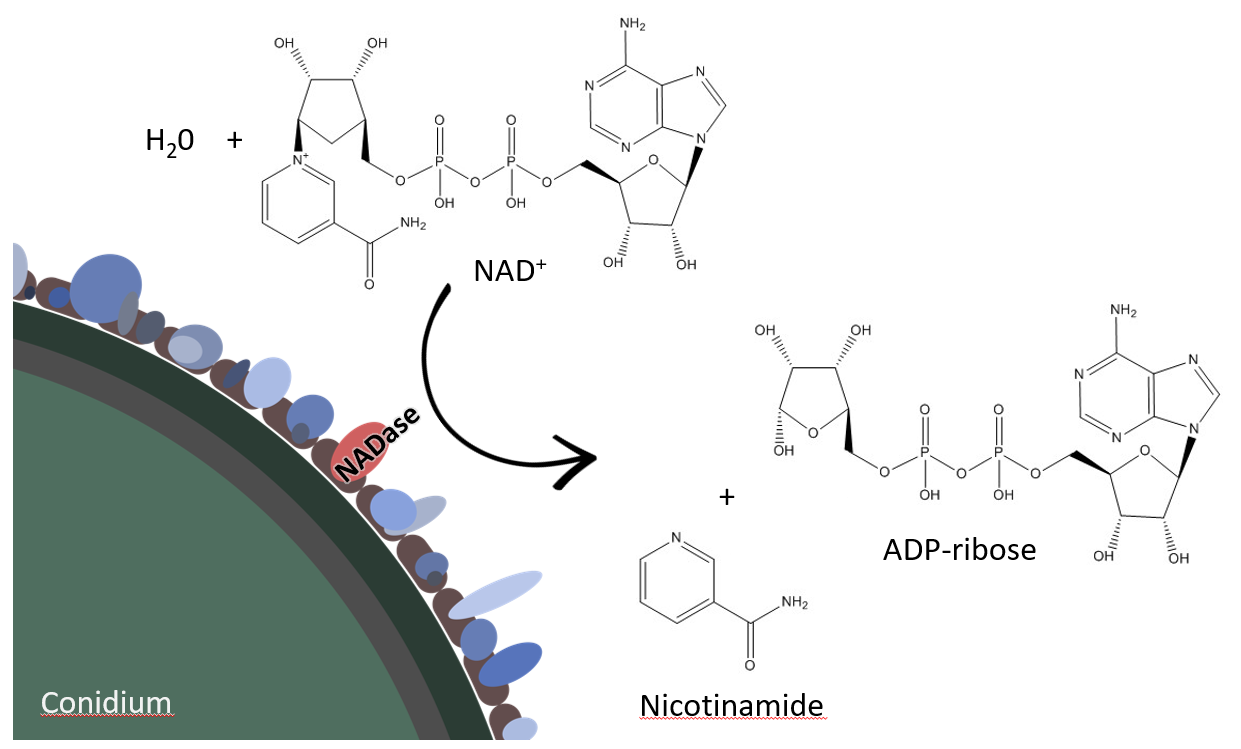
When we continued our work on the conidial surface proteome, the abundant surface protein Afu6g14470 attracted our attention, since it contains a tuberculosis necrotizing toxin domain (TNT). This domain is present in the protein CpnT of Mycobacterium tuberculosis and is known to induce necrosis of infected cells due to its NAD+ hydrolyzing activity. Coincidently, Mathias Ziegler, the head of the Department of Biomedicine of the University of Bergen, contacted us about a potential NADase activity on the spores of A. fumigatus. As a result, a very fruitful collaboration was born. By using enzyme activity gels, the Ziegler group together with us identified fragments of the protein sequence, which turned out to be protein Afu6g14470, the same NADase we had discovered on the spore surface. Øvind Strømland from the group of Mathias Ziegler produced the enzyme in insect cells (Sf9). By elegant biochemical work, the lab in Bergen was able to elucidate its structure and catalytic mechanism, which is distinct from known NADases. By gene deletion and further experimentation, we demonstrated that the A. fumigatus genome contains only a single gene encoding an NADase. The NADase activity is not only present on conidia, but also on hyphae and is detectable on different clinical strains independent of the culture conditions. We propose that the A. fumigatus NADase conveys an advantage during interaction with the host or competing microorganisms, but its exact biological role has yet to be revealed. So there is more to be uncovered.
References
- Stop neglecting fungi. (2017) NatMicrobiol 2: https://doi.org/10.1038/nmicrobiol.2017.120
- Latgé J.-P, Chamillos G. (2019) Aspergillus fumigatus and Aspergillosis in 2019. Clin Microbiol Rev 33:e00140-18.
- Blango MG, Kniemeyer O, Brakhage AA (2019) Conidial surface proteins at the interface of fungal infections. PLoS Pathog 15:e1007939.
- Voltersen V, Blango MG, Herrmann S, Schmidt F, Heinekamp T, Strassburger M, Krüger T, Bacher P, Lother J, Weiss E, Hünniger K, Liu H, Hortschansky P, Scheffold A, Löffler J, Krappmann S, Nietzsche S, Kurzai O, Einsele H, Kniemeyer O, Filler SG, Reichard U, Brakhage AA. (2018) Proteome analysis reveals the conidial surface protein CcpA essential for virulence of the pathogenic fungus Aspergillus fumigatus. mBio 9:e01557-18.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in