
Plants use multiple strategies to defend themselves, including a diverse defensive arsenal of hundreds of thousands of compounds. Among these are the isothiocyanates, the chemical defenses of cruciferous (Brassicales) plants that give mustards and radishes their spicy tang. While most of us love the flavors of these compounds, they are very effective antiherbivore and antimicrobial defenses, and help protect crucifer crops such as broccoli and cabbage vegetables against damage.
Still, there are herbivores and microbes that can attack crucifers with impunity, in spite of isothiocyanates. A handful of elegant biochemical mechanisms were found to be responsible for many of these cases, such as the conjugation of isothiocyanates with glutathione in several insects and in mammals (including humans). In some pathogenic bacteria, on the other hand, so-called Sax genes were discovered in 2011 (Fan et al., DOI: 10.1126/science.1199707), and were named for increasing their Survival in Arabidopsis eXtracts. However, whether phytopathogenic fungi also use biochemical detoxification to successfully attack isothiocyanate-producing plants was not well understood.
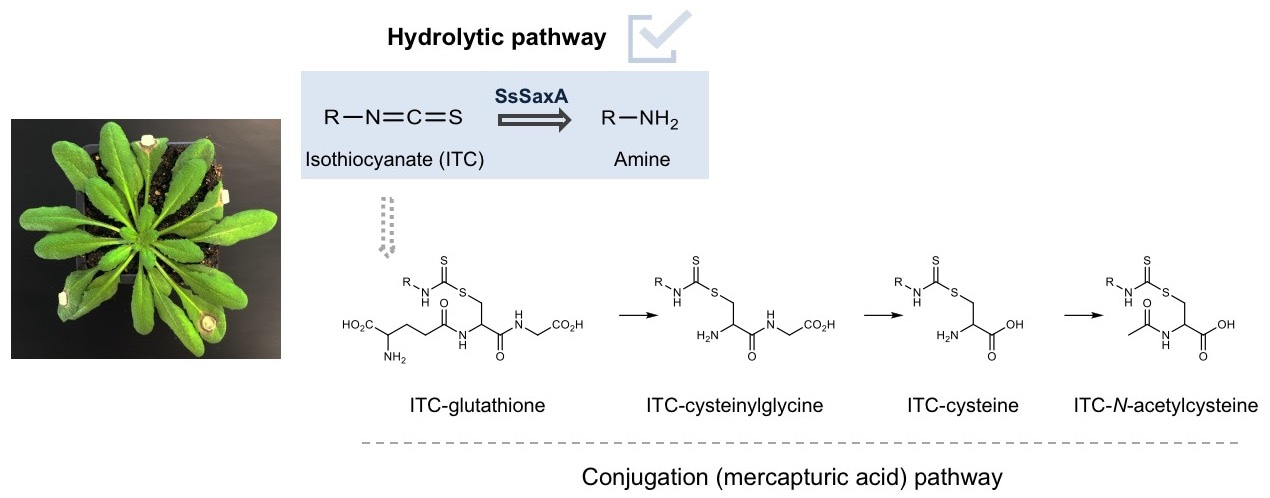
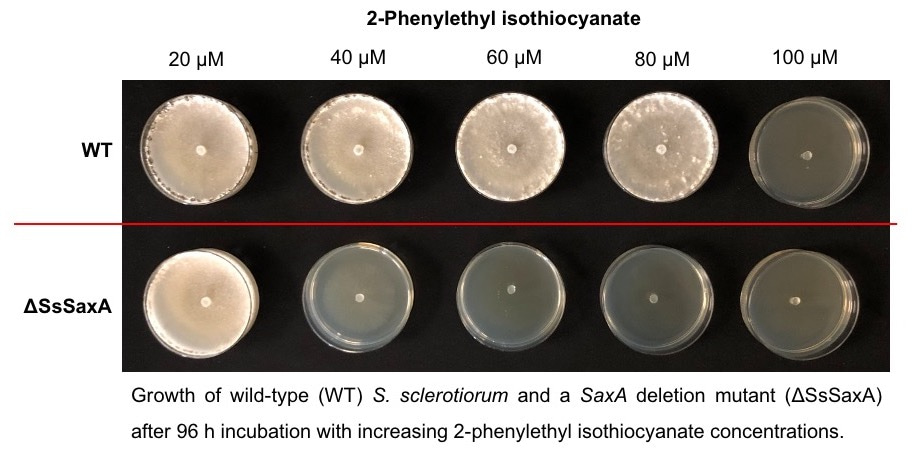
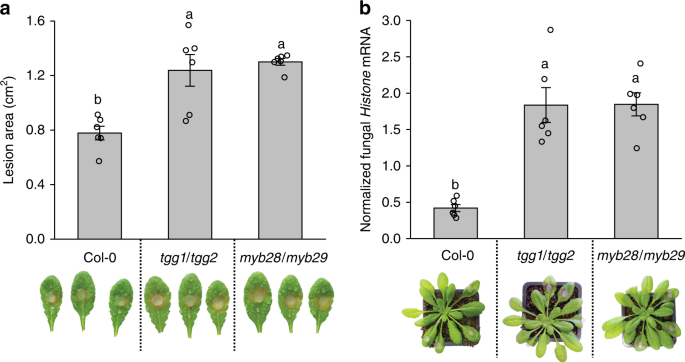
In this manuscript, we found that Sclerotinia sclerotiorum, a pathogenic fungus that causes white mold diseases in many plants all over the world, not only conjugates isothiocyanates to glutathione, but also hydrolyzes isothiocyanates to amines using a SaxA protein. This protein is an isothiocyanate hydrolase, and it degrades plant isothiocyanates into non-toxic amine derivatives. The metabolism of isothiocyanates via the latter degradation pathway is more than a hundred times more effective than via the general conjugation pathway. Consistently, lab-generated SaxA-deleted mutant fungi show significantly reduced tolerance to isothiocyanates in culture and in planta, even though the glutathione conjugation pathway was up-regulated by the mutants to compensate for the missing hydrolytic pathway.
Sequences of similar SaxA-like proteins are also present in the genomes of other phytopathogenic fungi, but haven’t yet been tested for activity, and their importance for plant disease is not yet known. But now that this reaction has been found in multiple kingdoms of life, it will be interesting to check whether it’s much more widespread and relevant than what was initially thought.
Jingyuan Chen, Almuth Hammerbacher and Daniel G. Vassão
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in