Gene Editing in Hematopoietic Stem Cells: One Step Forward
Published in Bioengineering & Biotechnology

Engineered nucleases, such as CRISPR/Cas, enable delivering double-stranded DNA breaks (DSBs) to a genomic site of choice, which may be used for target gene disruption when the break is sealed. Gene correction strategies, however, require homology-directed repair (HDR) of the DSB through an exogenous template (such as Adeno Associated Vectors, AAVs), which allows editing the target sequence as desired (Figure 1). Unfortunately, this process is highly inefficient in hematopoietic stem cells (HSCs), thus hindering its clinical translation1.
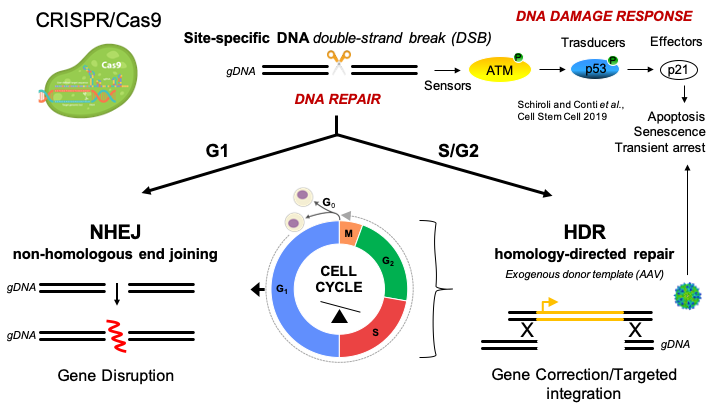
Figure 1. Editing-induced mechanisms in HSCs.
We developed a "3-days" genome editing protocol2-3 where HSCs are pre-stimulated ex-vivo to favor activation of the primitive cell subset4. On day 3 after thaw/isolation, the CRISPR/Cas9 nuclease is complexed with specific gRNAs for a targeted locus and delivered to HSCs by electroporation as pre-assembled ribonucleoprotein (RNP). HSCs are then transduced with an AAV6 carrying the HDR donor template. The electroporation step can be also exploited to transiently co-deliver mRNA(s) encoding for proteins of interest. Finally, treated cells require 24 hours for complete recovering before transplantation in immunodeficient NSG (NOD scid gamma mouse) mice. A small fraction of cells is kept in culture in order to perform deeper characterization (Figure 2).
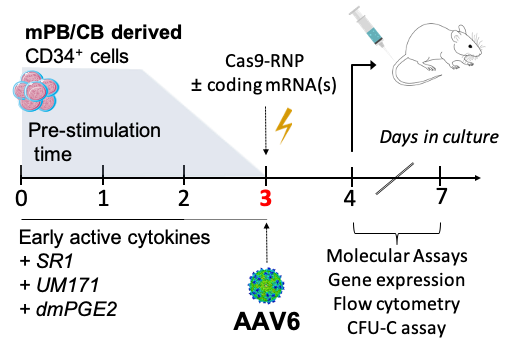
Figure 2. Ex-vivo HSC editing protocol.
We recently reported that HDR-mediated gene editing triggers robust p53 pathway activation in human HSCs, which reduces hematopoietic reconstitution in xenograft mouse models. Transient expression of a dominant negative p53 mutant protein (GSE56) during editing rescued the size of human graft5. However, it remained unknown whether such outcome was due to altered growth properties rather than improved preservation of repopulating edited HSCs. Indeed, little information is available about clonogenic output and multilineage repopulation of human edited HSCs, even in absence of p53 inhibition.
We were concerned about oligoclonal hematopoiesis of treated cells which may delay hematopoietic recovery, impair graft resilience and potentially expose patients to higher risk of leukemia and myelodysplastic syndrome6. Yet, the reconstitution of an identical sequence at the targeted site in HDR-edited clones was the major technical challenge hindering clonal tracking of edited HSCs, thus prompting the need for ad hoc strategies. We thus generated a highly complex library of AAV donor templates tagged with unique hereditable barcodes (Figure 3). By using such clonal tracking approach (BAR-Seq), we highlighted the significant “cost” of the editing procedure, which substantially lowers the number of clones engrafting in recipient mice. We were surprised how transient inhibition of the editing-induced p53 response rescues polyclonal composition of the human edited cells graft, without altering size and dynamic of the individual clonal output.
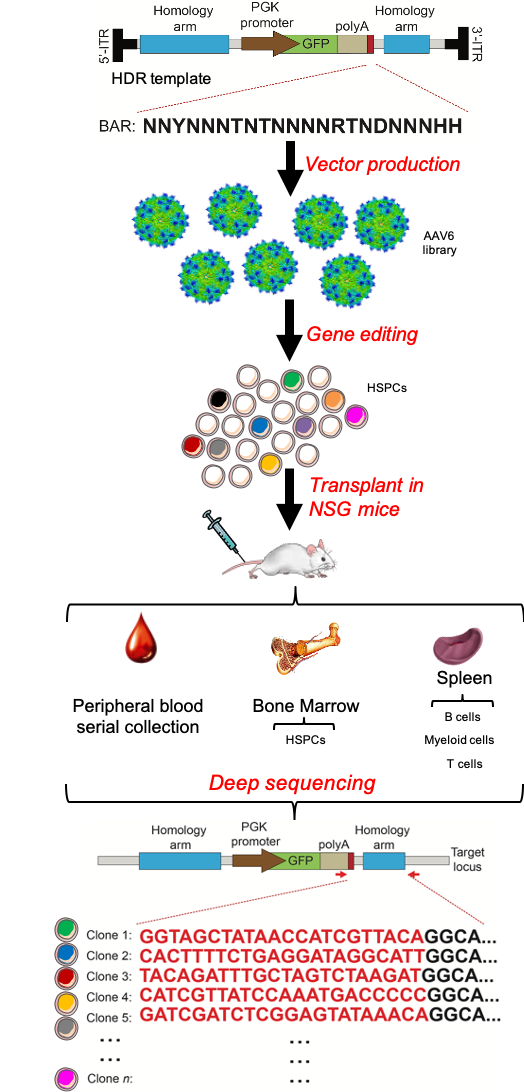
Figure 3. Pipeline of BAR-Seq technology.
Beside preserving polyclonality, therapeutic HSC products require high proportion of corrected cells for clinical benefit. Cell quiescence, delayed cell-cycle progression and low expression of the DNA DSB repair machinery were the major biological challenges to proficient HDR-editing, although the underlying mechanisms of such blocks remained unclear. Our work highlights that pervasive upregulation of the cellular HDR machinery accompanying engagement in S/G2 phases is a crucial factor enabling efficient HDR-editing in HSCs. Intriguingly, such complex choreography can be set in motion by transient expression of an adenoviral protein (Ad5-E4orf6/7) naturally evolved to plug directly into the master cell cycle regulator E2F7 (Figure 4). By combining transient expression of this newly found adenoviral effector with the p53 inhibitor, we reached high and stable proportions of HDR-edited HSCs in long-term human xenografts (up to 50%), which surpass those reported in literature until now.
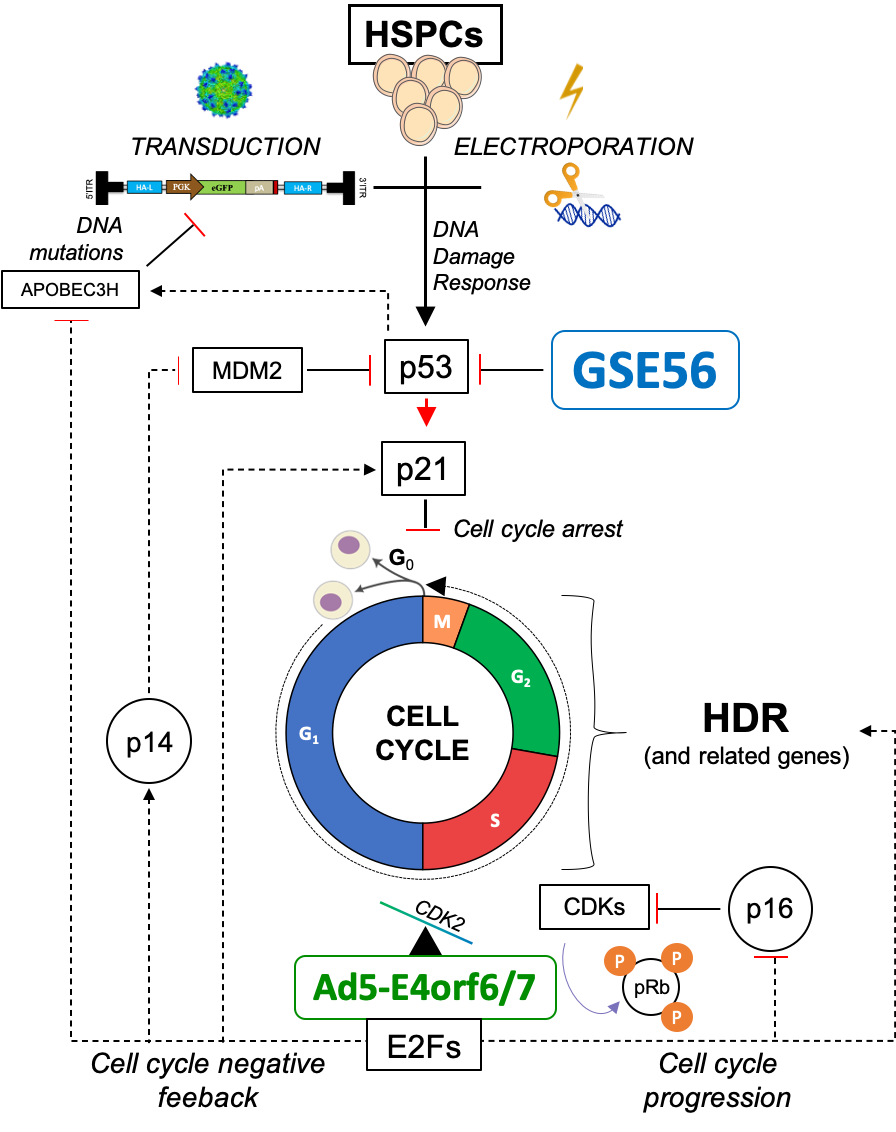
Figure 4. Molecular mechanisms engaged upon enhanced editing.
Importantly, BAR-Seq stringently validated our enhanced editing strategy and showed full preservation of in-vivo clonogenic output, multipotency and self-renewal capacity of edited HSCs over serial transplantation. We envisage that the gains obtained by the enhanced editing in clonal repertoire and percentage of edited cells are highly relevant for clinical translation (Figure 5). We thus expect that our enhanced protocol will allow safer and more effective clinical application of HSC gene editing.
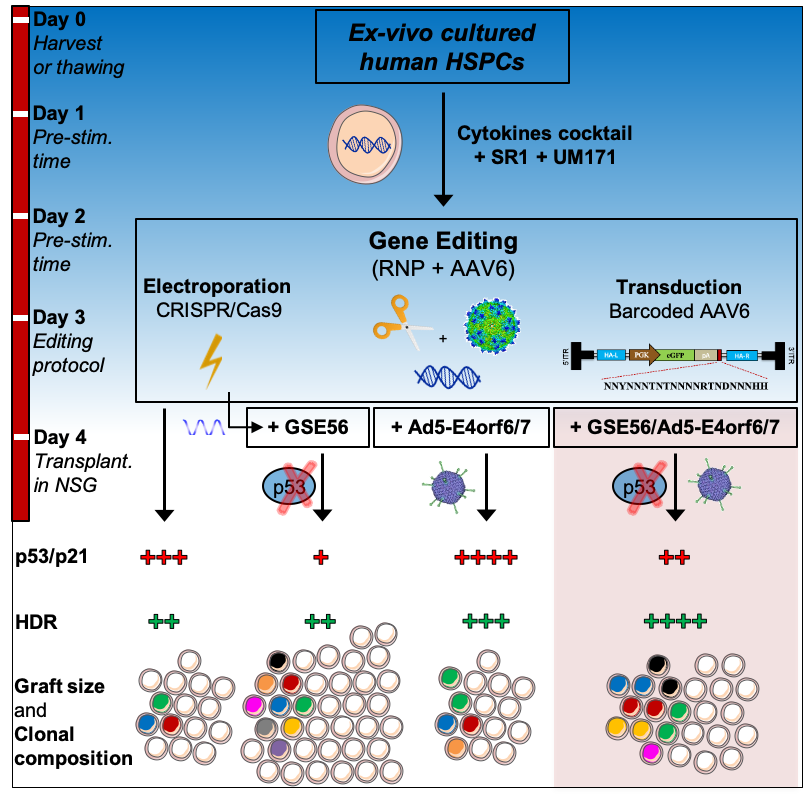
Figure 5. Summary of HSC editing strategies and their outcomes.
To know more about how we moved HSC gene editing one step forward, please download the full article: Efficient gene editing of human long-term hematopoietic stem cells validated by clonal tracking
REFERENCES
3. Schiroli, G. et al. Schiroli, G. et al. Preclinical modeling highlights the therapeutic potential of hematopoietic stem cell gene editing for correction of SCID-X1. Sci. Transl. Med. 9, (2017).
7. Huang, M. M. & Hearing, P. The adenovirus early region 4 open reading frame 6/7 protein regulates the DNA binding activity of the cellular transcription factor, E2F, through a direct complex. Genes Dev. 3, 1699–1710 (1989).
Follow the Topic
-
Nature Biotechnology

A monthly journal covering the science and business of biotechnology, with new concepts in technology/methodology of relevance to the biological, biomedical, agricultural and environmental sciences.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in