Generating synthetic mixed-type longitudinal electronic health records for artificial intelligent applications
Published in Healthcare & Nursing
What led to this study
Machine learning-based digital health solutions have been increasingly applied to advance health- care. The exponential growth of medical data, such as electronic health records (EHRs) from healthcare systems, has facilitated the development of novel methodologies empowered by artificial intelligence (AI). However, gaining access to patient data in healthcare organizations can be quite a hurdle. Due to significant concerns regarding patient privacy, stakeholders are often reluctant to share medical data. Consequently, a dearth of available data for biomedical research arises, impeding the progress of machine learning in computational health.
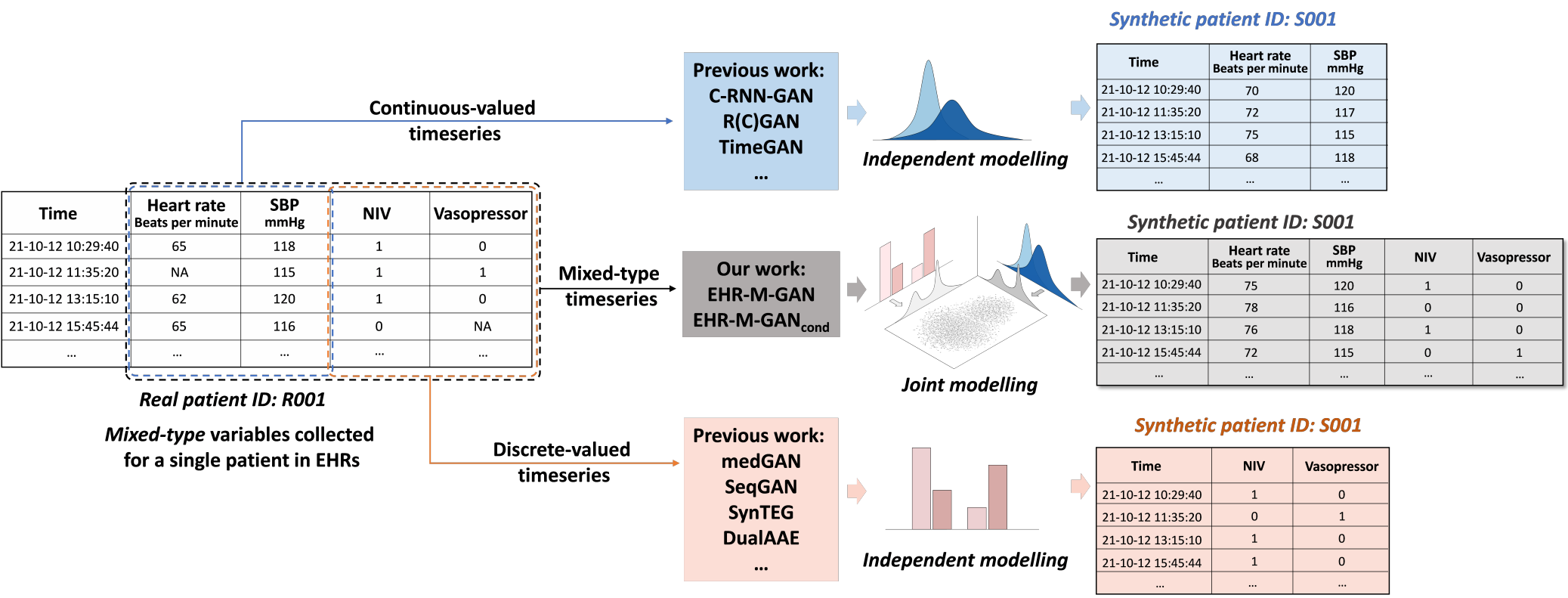 Figure 1. Comparison between different generative methods. Models in the previous literature are mostly limited in generating single type of clinical data. The proposed model can simultaneously synthesize mixed-type timeseries while modeling their correlations.
Figure 1. Comparison between different generative methods. Models in the previous literature are mostly limited in generating single type of clinical data. The proposed model can simultaneously synthesize mixed-type timeseries while modeling their correlations.
One viable solution is for the data holder to create synthetic data. By retaining the statistical characteristics of the original data, synthetic data can ensure its representativeness and utility for downstream analysis while avoiding privacy breaches. This approach of synthesizing data has demonstrated successful applications in various projects, including the Synthea [1], Clinical Practice Research Datalink (CPRD) [2], and the National COVID Cohort Collaborative (N3C) during the COVID-19 pandemic [3].
However, the task of synthesizing realistic EHRs can be tricky, especially for synthesizing longitudinal health records which contain patient trajectories. First, EHRs encompass heterogeneous data types, as clinical decision-making in real practice includes a variety of information sources. For example, patient physiological signals and laboratory test results are collected in the EHR as continuous-valued timeseries, while the medication and diagnostic information are recorded as discretized-valued data, such as categorical ICD codes (see Figure 1). Moreover, although mixed-type clinical timeseries are heterogeneous regarding the data types, they are highly correlated and inform one another of the underlying health of an individual. It is therefore hard to simultaneously synthesizing mixed-type EHR timeseries that differ in syntax and distributions, while preserving the temporal correlations between them (see Figure 2a).
What we did
To address this, we propose a GAN framework called EHR-M-GAN (see Figure 2b). The proposed model can synthesize patient trajectories with high-dimensionality and heterogeneous data types (both continuous-valued and discrete-valued timeseries), while capturing the underlying temporal dependencies.
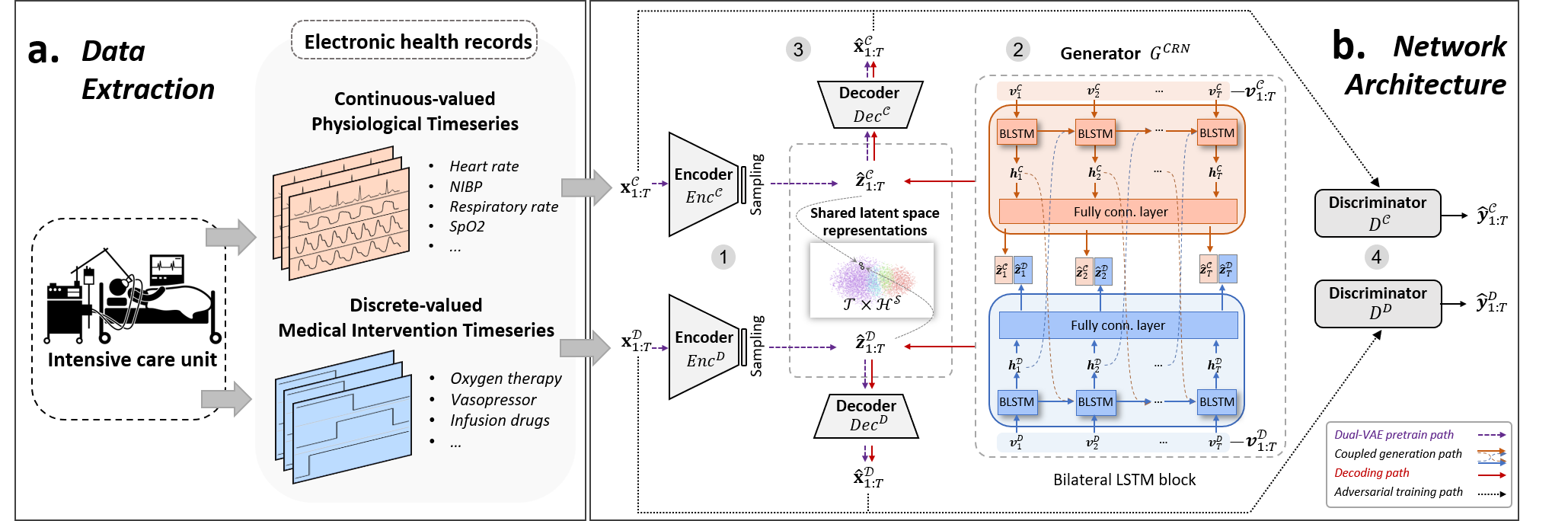
Figure 2. a. Data extraction. EHRs with mixed data types are routinely collected for patients in intensive care units (ICUs). b. Network architecture. The proposed model, EHR-M-GAN, contains two key components — dual variational autoencoder and sequentially coupled generator.
Specifically, we first jointly model the underlying distributions of the heterogeneous features by introducing dual variational autoencoder (dual-VAE). By incorporating multiple loss constraints, including matching loss and contrastive loss, the distribution gap can be effectively mitigated through the utilization of lower-dimensional shared latent representations. Then, to capture the correlated temporal dynamics of the mixed-type timeseries, a sequentially coupled generator is employed in the networks of generative adversarial networks (GANs) (see Fig. 2b).
What we found
The validation of the proposed model is based on critical care setting, where the intensive care units (ICU) patients are closely monitored with mixed-type EHR timeseries collected. Evaluations are performed based on three publicly available ICU datasets — MIMIC-III [4], eICU [5] and HiRID [6] from a total of 141,488 patients. Our EHR-M-GAN outperforms the state-of-the-art benchmarks on a diverse spectrum of evaluation metrics, with regard to fidelity, correlation, utility, and privacy of the synthetic data (see Table 1 for the evaluation pipeline).
Table 1. Evaluation protocol in this study.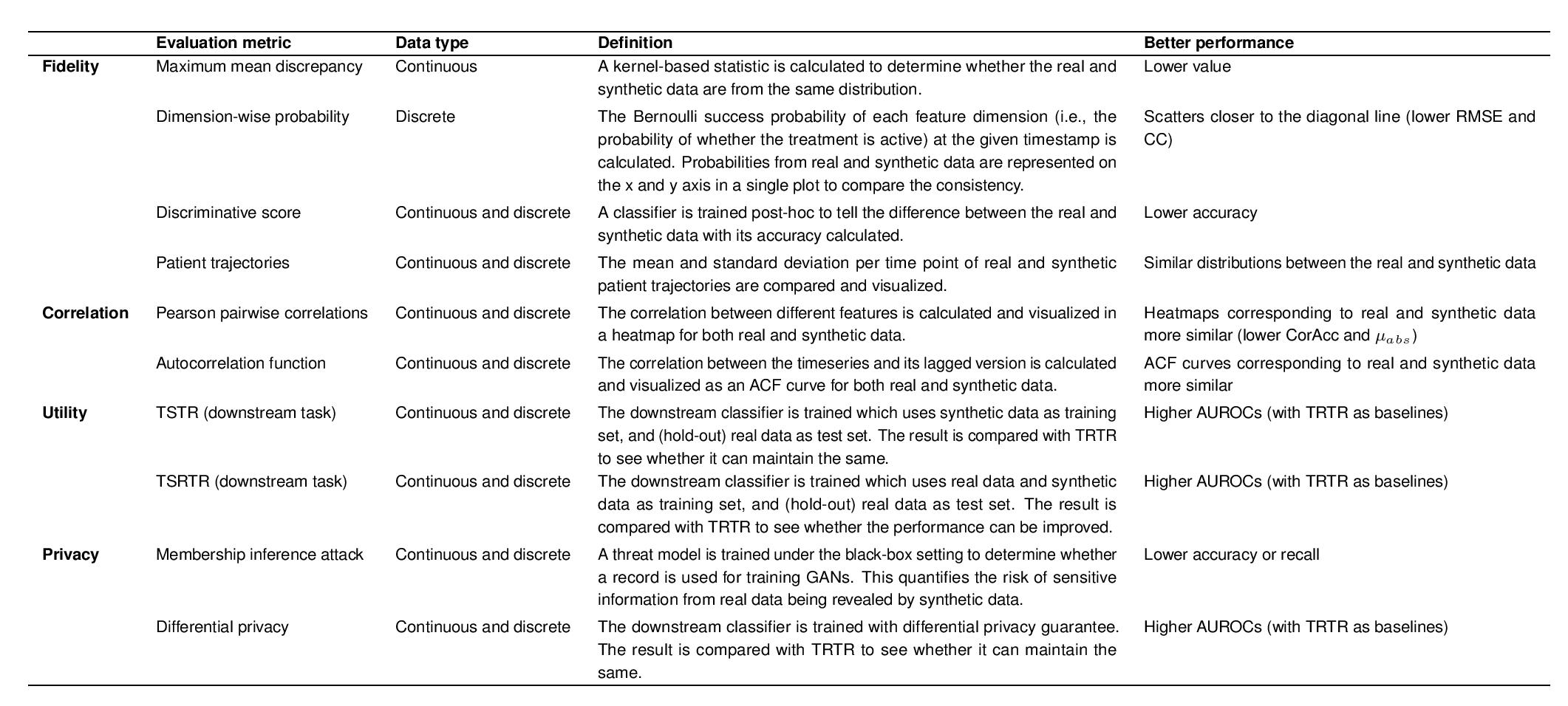
Notably, the experimental results indicate that the quality of the synthetic EHR data can be improved by the integration of mixed-type information, in contrast to the benchmarks that utilize single-type data for learning. In the ablation study, our proposed model also outperforms other GAN-based variants that allow mixed-type inputs, indicating that the components in the proposed model are effective in synthesizing mixed-type timeseries with high fidelity, while successfully reconstructing the interdependencies between them (see Figure 3). During downstream task evaluation, given the prediction of medical interventions in fast-paced critical care environments as an exemplar, the results demonstrate that the robustness of machine learning algorithms can be improved by data augmentation based on the synthetic EHR timeseries (see Table 2).
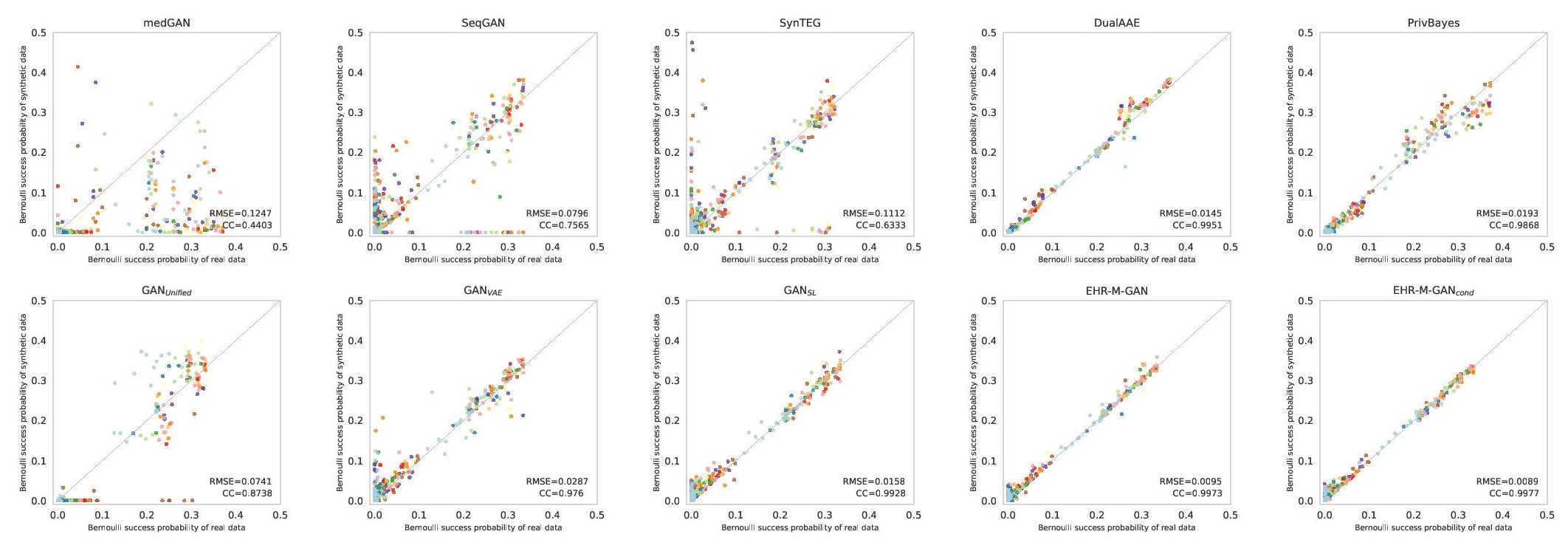
Figure 3. Scatterplot of the dimension-wise probability test on MIMIC-III dataset. The x-axis and y-axis represent dimension-wise probability for the real data and synthetic data generated from different models, respectively. The optimal performance appears along the diagonal line.
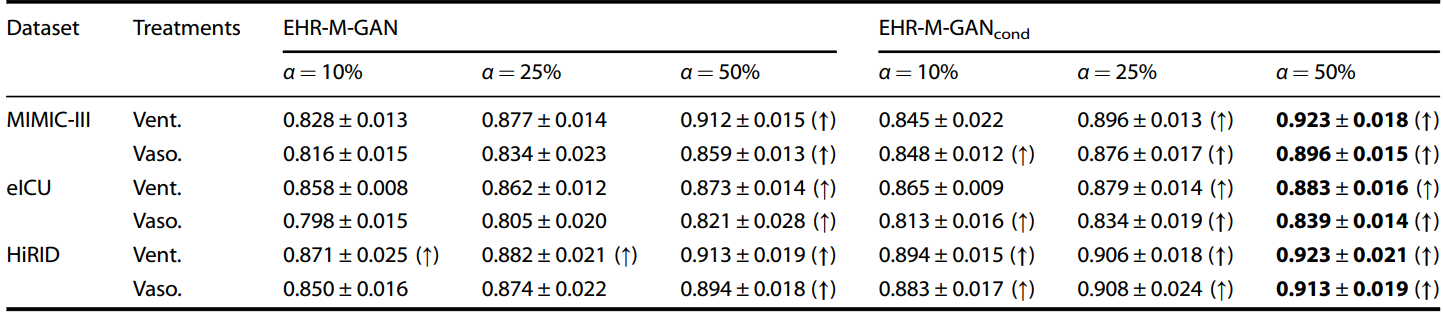
Table 2. Downstream task evaluation with data augmentation ratio α using the synthetic EHR timeseries. The upper arrow indicates that the AUROC value under Train on Synthetic and Real, Test on Real (TSRTR) is higher than Train on Synthetic, Test on Real (TRTR) for the corresponding task. The bold arrow indicates that the value is significantly improved using t-test (p ≤ 0.05). Bolded values denote best scores. The detailed experimental settings can be found in the original paper.
What it means
The synthetic EHR data circumvent the issue of privacy breaches, therefore offering a viable solution to overcome the challenges posed by the scarcity of medical data available for research purposes. With the utility of the original EHR data being well-preserved, the synthetic data enables healthcare professionals to implement cutting-edge machine learning algorithms, create benchmarks, and foster collaboration through data sharing.
By synthesizing various types of EHR data and better mimicking the nature of clinical decision-making, the effectiveness and usability of the downstream AI applications can be enhanced. It is now feasible to use the synthesized data to improve the performance of machine learning models for downstream applications such as the prediction of the next intervention, or patient phenotyping, based on both the continuous and discrete components of EHR timeseries. Therefore, the proposed EHR synthesizer has the potential to expedite the development of AI-powered clinical tools in real-world healthcare settings.
References
[1] https://synthetichealth.github.io/synthea/
[2] https://cprd.com/synthetic-data
[3] https://ncats.nih.gov/n3c/
[4] Johnson A E W, Pollard T J, Shen L, et al. MIMIC-III, a freely accessible critical care database[J]. Scientific data, 2016, 3(1): 1-9.
[5] Pollard T J, Johnson A E W, Raffa J D, et al. The eICU Collaborative Research Database, a freely available multi-center database for critical care research[J]. Scientific data, 2018, 5(1): 1-13.
[6] Yèche H, Kuznetsova R, Zimmermann M, et al. HiRID-ICU-Benchmark--A Comprehensive Machine Learning Benchmark on High-resolution ICU Data[J]. arXiv preprint arXiv:2111.08536, 2021.
Follow the Topic
-
npj Digital Medicine

An online open-access journal dedicated to publishing research in all aspects of digital medicine, including the clinical application and implementation of digital and mobile technologies, virtual healthcare, and novel applications of artificial intelligence and informatics.
Related Collections
With Collections, you can get published faster and increase your visibility.
Evaluating the Real-World Clinical Performance of AI
Publishing Model: Open Access
Deadline: Jun 03, 2026
Impact of Agentic AI on Care Delivery
Publishing Model: Open Access
Deadline: Jul 12, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in