Generation of CRISPR/Cas9-mediated genetic knockout human intestinal tissue-derived enteroid lines by lentivirus transduction and single cell cloning
Published in Protocols & Methods

In 2011, there was a breakthrough in the gastrointestinal (GI) tract research field by Sato and Clevers, who established tissue-derived human intestinal enteroids (HIEs, also called organoids or miniguts) to mimic the human GI tract epithelium as an ex vivo culture model (1). The HIEs are nontransformed epithelial cultures derived from intestinal stem cells, which retain the capacity for self-renewal and stem cell pluripotency during development. With those stem cells still present in the cultures, HIEs are able to grow indefinitely, and after a removal of growth factors from the medium used to maintain the cultures, the stem cells then differentiate into multicellular epithelial cultures with enterocytes, enteroendocrine cells, goblet cells and Paneth cells present as in the human GI tract epithelium in vivo (1). Human biopsy-derived HIEs are a powerful tool for human host-oriented translational research and precision medicine, since these cultures recapitulate host genome variability, intestine segment specificity and are physiologically active, making them relevant ex vivo models. Many different groups have used HIEs in GI, stem cell biology, cancer, virology, bacteriology, and parasitology research for cultivating human trophic microbes including previously noncultivatable microorganisms (2-11).
Notably, in 2016, HIEs were used as the first ex vivo intestinal cultivation model for human norovirus (HuNoV) (11), which was noncultivatable in all available cell lines since discovery of the virus in 1968. Excitingly, the HuNoV-HIE model supports reproducible replication of multiple HuNoV strains, and confirms epidemiological differences in genetic susceptibility in people infected with HuNoV (8, 12, 13). Moreover, HuNoV replicates in the intestinal enterocytes of HIEs, which recapitulates the tissue tropism observed in human patients. The model also illustrates strain-specific requirements for HuNoV replication including the requirement of genetically controlled histo-blood group antigens (HBGAs) for GII.4 HuNoV infection and replication, and the requirement of bile acids for GII.3 HuNoV replication (14, 15). As a result, the HIE culture system allows new studies on virology, gut microbiology, GI research, host-pathogen interactions, antibody-mediated neutralization and other antiviral interventions for controlling pathogen infections.
Originally, the CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 endonuclease was identified as a component of the CRISPR Escherichea coli bacterial innate immunity system (16). The Cas9 gene encodes a novel class of sequence-specific endonuclease complexes to eliminate foreign DNA from bacteriophages. Interestingly, the target specificity of the Cas9 endonuclease is solely determined by its accompanying small-guiding RNA (sgRNA) sequence (17-20) rather than the protein motifs. Therefore, Cas9 can be easily reprogrammed to target a desired sequence in the host genome, by replacing that 20-bp sgRNA sequence for specificity determination. This simple property makes Cas9 superior to other endonuclease systems and to be the most well-known and easy-to-use designer nuclease technique for all biological research fields. The only requirement of the target site is that it must be followed by the protospacer adjacent motif (PAM) as “NGG” (N for any nucleotide) for Cas9 recognition (17).
After recognizing the sequence, the active Cas9 endonuclease can generate a double-strand break (DSB) at the target site to activate the host endogenous DNA repair system (21, 22). In the absence of a homologous template, the DSB can be repaired by joining the two DNA ends together, called non-homologous end-joining (NHEJ) (23), which has a chance to result in deletion or insertion of base-pairs (indels), producing frameshift mutations to truncate the target gene by generating early stop codons (24). Importantly, the target gene will acquire a loss-of-function phenotype or be knocked out if the frameshift-induced truncation occurs in its both alleles in the host genome. Before Cas9 was introduced, the DSB could only be generated by chance or by engineering the protein motifs on designed zinc-finger nucleases (ZFNs) or transcription activator-like effector nuclease (TALENs), which are harder and more time-consuming to construct than by simply ordering new primers to change the sgRNA sequence using the Cas9 system. As a result, the rapid and flexible Cas9 technique quickly dominated the fields of genome editing and genetics research.
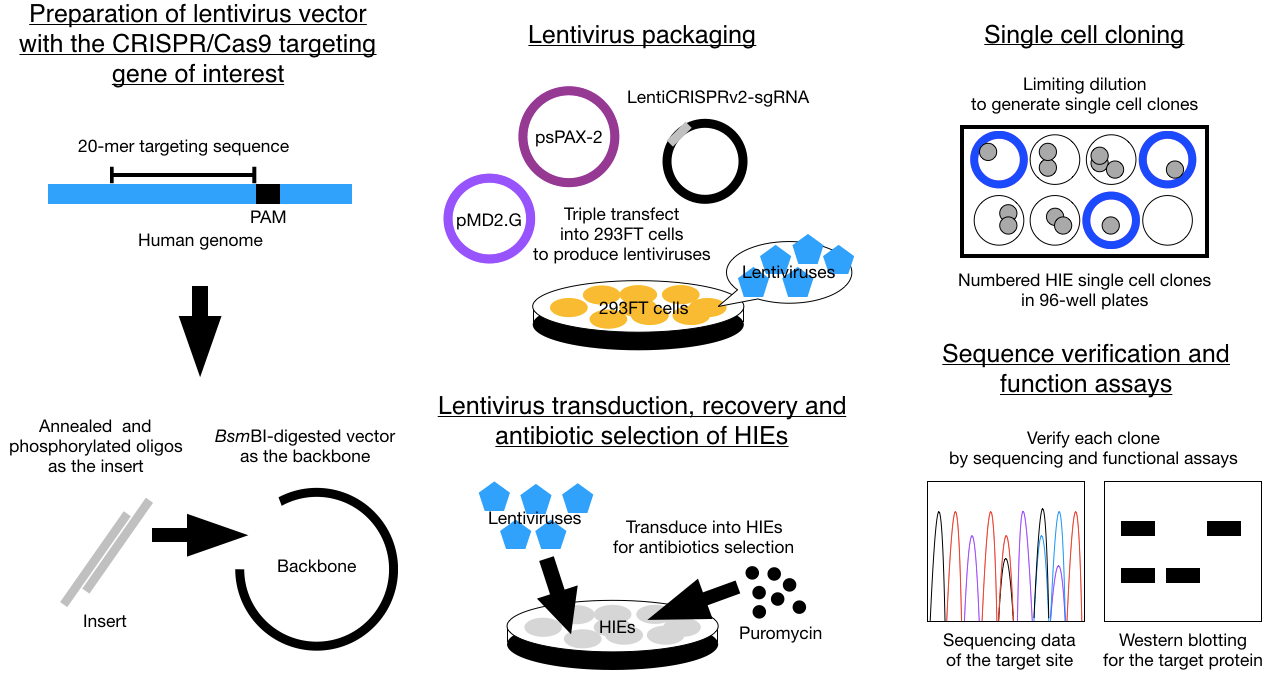
This protocol is an updated version of the previous Cas9-mediated gene knockout (KO) method developed and used by our lab in two publications in 2020 (12, 14). For the CRISPR system, we started from the traditional LentiCRISPRv2 system established by the Zhang lab. in MIT (25), merged with a 2nd generation system by using pMD2.G and pSPAX2 plasmids established by the Trono Lab in EPFL, Switzerland for lentivirus packaging. Although several articles have been published with the introduction of Cas9 into organoids, they are either technically difficult or use complicated cloning techniques. When we started our own trials, we found either electroporation or overnight transduction in 3D cultures were not optimal for Cas9 delivery into HIEs. Thus, we switched to lentiviral transduction, accompanied with an extra step by dispersing HIEs into single cells with trypsin-EDTA for spin inoculation, for gene delivery. This modification led to our final protocol outlined in two recent publications (12, 14).
The CRISPR/Cas9 technique has a lot of potential in HIE research, since it allows a scientist the opportunity to identify critical genes that affect stem cell differentiation, host-pathogen interactions, viral-induced innate immune responses and intestinal biology in many different fields. With this protocol, we successfully knocked out genes that affect HuNoV infection, including a host attachment factor gene FUT2 and an innate immunity gene STAT1 (12, 14). We and others have also used KO lines to investigate rotavirus infection and identified new mechanisms by which calcium waves induced by infection affect neighboring cells in KO lines (26, 27). Moreover, we can easily confirm the specificity of antiviral drug treatments that target host functions by characterizing affected downstream cellular signaling pathways in the KO HIE cultures. Beside being useful to study virus-host interactions, organoids that contain a specific gene KO can be used to identify mechanisms of pathogen entry or egress, signaling pathways, nutrient uptake and cellular metabolic processes including lipid metabolism, energy production, phospholipid biogenesis and pathophysiological consequences with different genes of interest and functional assays.
In summary, we are excited to share this simple protocol for making KO HIEs by providing step-by-step instructions for lentiviral transduction and single cell cloning. The technical requirements of knocking out a gene in HIEs are higher than in common cell lines since HIEs are more fragile, slow-growing and sensitive to environmental changes. Hopefully more labs can adapt the Cas9 technique in different organoid cultures easily with the help of this protocol.
Shih-Ching Lin, Kei Haga, Xi-Lei Zeng & Mary K. Estes
References
- T. Sato et al., Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 141, 1762-1772 (2011).
- I. Heo et al., Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat Microbiol 3, 814-823 (2018).
- S. Middendorp et al., Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells 32, 1083-1091 (2014).
- J. Foulke-Abel et al., Human enteroids as an ex-vivo model of host-pathogen interactions in the gastrointestinal tract. Exp Biol Med (Maywood) 239, 1124-1134 (2014).
- A. Rajan et al., Novel segment- and host-specific patterns of enteroaggregative Escherichia coli adherence to human intestinal enteroids. mBio 9 (2018).
- J. G. In et al., Human mini-guts: new insights into intestinal physiology and host-pathogen interactions. Nat Rev Gastroenterol Hepatol 13, 633-642 (2016).
- N. C. Zachos et al., Human enteroids/colonoids and intestinal organoids functionally recapitulate normal intestinal physiology and pathophysiology. J Biol Chem 291, 3759-3766 (2016).
- H. Clevers, COVID-19: organoids go viral. Nat Rev Mol Cell Biol 21, 355-356 (2020).
- M. H. Geurts, J. van der Vaart, J. Beumer, H. Clevers, The Organoid Platform: Promises and Challenges as Tools in the Fight against COVID-19. Stem Cell Reports 16, 412-418 (2021).
- M. F. Hares et al., Stem cell-derived enteroid cultures as a tool for dissecting host-parasite interactions in the small intestinal epithelium. Parasite Immunol 43, e12765 (2021).
- K. Ettayebi et al., Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387-1393 (2016).
- S. C. Lin et al., Human norovirus exhibits strain-specific sensitivity to host interferon pathways in human intestinal enteroids. Proc Natl Acad Sci U S A 117, 23782-23793 (2020).
- K. Ettayebi et al., New Insights and Enhanced Human Norovirus Cultivation in Human Intestinal Enteroids. mSphere 6(2021).
- K. Haga et al., Genetic manipulation of human intestinal enteroids demonstrates the necessity of a functional fucosyltransferase 2 gene for secretor-dependent human norovirus infection. mBio 11 (2020).
- K. Murakami et al., Bile acids and ceramide overcome the entry restriction for GII.3 human norovirus replication in human intestinal enteroids. Proc Natl Acad Sci U S A 117, 1700-1710 (2020).
- Y. Ishino, H. Shinagawa, K. Makino, M. Amemura, A. Nakata, Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. Journal of bacteriology 169, 5429-5433 (1987).
- M. Jinek et al., A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, N.Y.) 337, 816-821 (2012).
- P. Mali et al., CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature biotechnology 31, 833-838 (2013).
- L. Cong et al., Multiplex genome engineering using CRISPR/Cas systems. Science (New York, N.Y.) 339, 819-823 (2013).
- J. A. Doudna, E. Charpentier, Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science346, 1258096 (2014).
- F. Paques, J. E. Haber, Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiology and molecular biology reviews : MMBR 63, 349-404 (1999).
- F. D. Urnov, E. J. Rebar, M. C. Holmes, H. S. Zhang, P. D. Gregory, Genome editing with engineered zinc finger nucleases. Nature reviews. Genetics 11, 636-646 (2010).
- J. K. Moore, J. E. Haber, Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Molecular and cellular biology 16, 2164-2173 (1996).
- S. Durai et al., Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic acids research 33, 5978-5990 (2005).
- O. Shalem et al., Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84-87 (2014).
- K. Saxena et al., Human Intestinal Enteroids: a New Model To Study Human Rotavirus Infection, Host Restriction, and Pathophysiology. J Virol 90, 43-56 (2016).
- A. L. Chang-Graham et al., Rotavirus induces intercellular calcium waves through ADP signaling. Science 370 (2020).
Follow the Topic
-
Nature Protocols

This journal publishes secondary research articles and covers new techniques and technologies, as well as established methods, used in all fields of the biological, chemical and clinical sciences.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in