
The mysterious gut microbe that does it all
The relationship between gut microbiome and health is well established, but there is still much to learn about the specific bacteria that reside in this environment. The gut microbiome contains a vast number of genes that encode proteins of unknown function. Unfortunately, many gut microbes can be difficult to cultivate and manipulate genetically, making it challenging to assign phenotypes to these genes.
Akkermansia muciniphila is a prominent gut microbe that lacked any methods for genetic manipulation. This bacterium colonizes the intestine, where it forages on components of the mucus layer called mucins. Initially isolated in the early 2000s by Muriel Derrien et al.1, interest in A. muciniphila has exploded recently as we learn more about its impact on health. Colonization with A. muciniphila has been shown to protect against obesity, infection, and neurological disorders, and to enhance the efficacy of cancer immunotherapy2–6. Despite these exciting links to human health, we still know very little about how A. muciniphila colonizes the gut and impacts its host. A major barrier to investigating these questions was a lack of genetic tools.
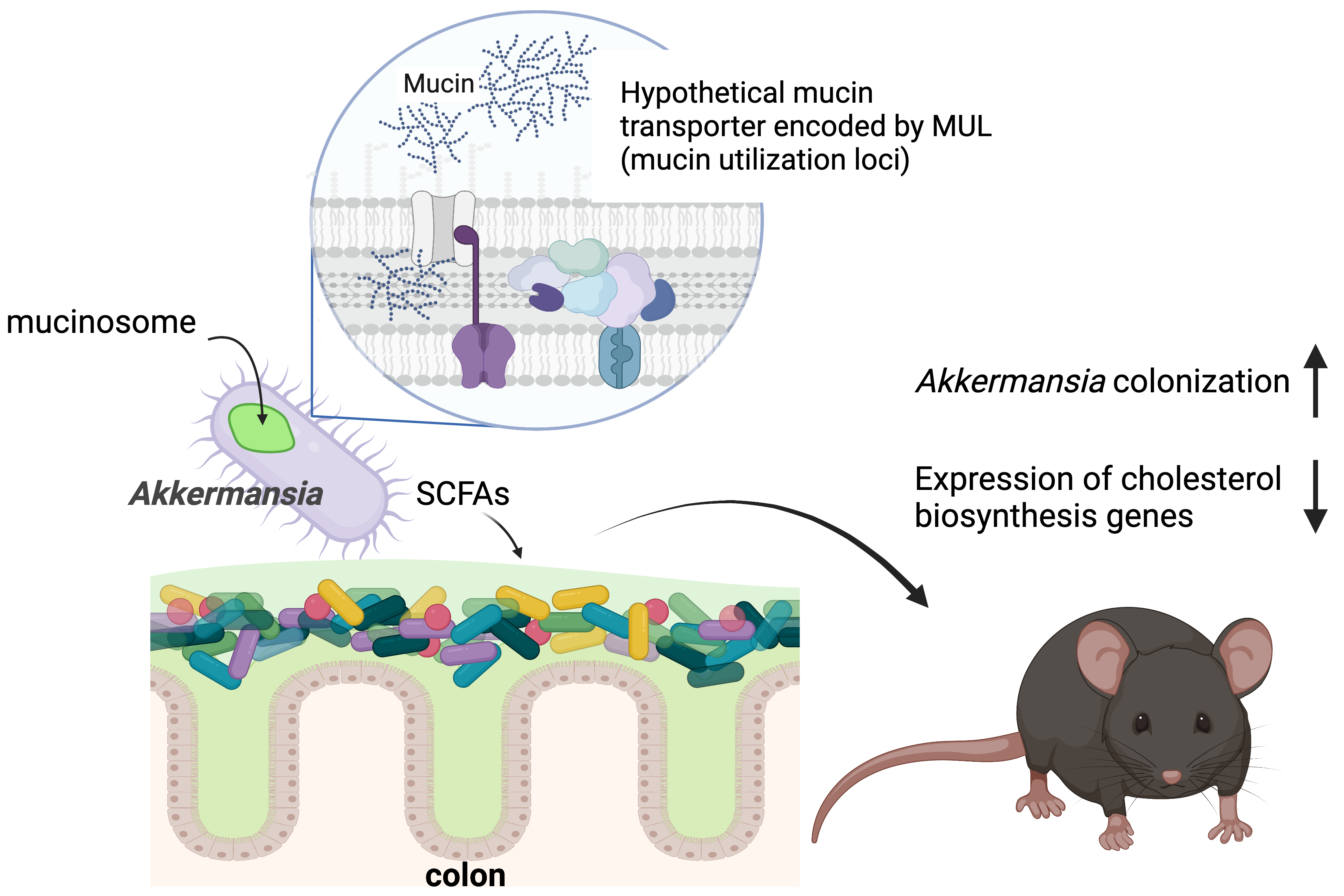
Using newly developed genetic approaches, we discovered a transporter required for A. muciniphila to import mucin into the cell, where it accumulates in mucinosomes. Mucin degradation affects short chain fatty acid (SFCA) production by A. muciniphila and alters the expression of lipid metabolism genes in the host.
Discovery of genes required for growth on mucin and a new subcellular structure, the "mucinosome"
In our recent Nature Microbiology paper7, we established methods for transposon mutagenesis in A. muciniphila, providing the first genetic clues to how this organism colonizes its niche in the gut. Using a high throughput screening technique called transposon insertion sequencing (INSeq/TnSeq), we identified genes required for A. muciniphila to grow on mucins and colonize the intestinal tract of various mouse models. Genes needed for A. muciniphila to survive in the gut were predicted to encode functions such as amino acid biosynthesis, secretion systems, and surface proteins. The mutations that caused the greatest growth defects, however, were in genes that had no predicted functions and lacked homology to any other organisms. We zeroed in on two clusters of these genes of unknown function that were essential for growth in mucin and named them Mucin Utilization Loci (MULs).
As its name suggests, A. muciniphila loves mucin. While other gut microbes can degrade mucins to varying extents, A. muciniphila is the only gut microbe known to specialize in using it as its preferred food source. Mucins are complex glycoproteins that are typically challenging for bacteria to degrade, raising the question of how A. muciniphila can use it so efficiently. To watch mucin uptake in real time, we fed A. muciniphila fluorescently labelled mucin. Unexpectedly, we found that it imported mucins and accumulated them in intracellular structures that we named mucinosomes. We hypothesize that this form of selfish eating that allows A. muciniphila to feed on mucins while limiting the amount of partially degraded glycans that could be lost to competing members of the microbiota. The MUL genes were essential for mucin transport and mucinosome formation. Without mutagenesis and phenotypic analysis, there were few clues to the functions of these genes.
Akkermansia's ability to eat mucin modulates cholesterol metabolism genes in the gut
By generating mutants that could not use mucin, we were in a position to investigate how mucin foraging by A. muciniphila impacts the host. We performed transcriptional analysis of germ-free mice colonized with wild-type A. muciniphila and MUL mutants. We found that mucin utilization by A. muciniphila repressed cholesterol biosynthesis genes in the colon. Our findings are consistent with previous observations that A. muciniphila and other gut microbes can modulate cholesterol metabolism8 and demonstrate that mucin eating activity in the gut is important for this function.
Future implications
Akkermansia has great potential for use as a probiotic, yet the factors that influence colonization and host-microbe interactions are not fully understood. By exploring the mechanisms underlying these interactions, we will be in a better position to leverage the therapeutic properties of this unusual bacterium.
Behind the Paper
The Valdivia lab has a history of pioneering methods for genetically manipulating fastidious microbes. The lab is a leader in Chlamydia genetics, an obligate intracellular pathogen and phylogenetic relative of Akkermansia. By combining chemical mutagenesis with high throughput whole genome sequencing, the lab has discovered mechanisms of pathogenesis in Chlamydia trachomatis, and flagellar components in another intractable microbe, Exiguobacterium acetylicum9,10. From here, the lab sought to tackle a member of the human gut microbiota and set its sights on Akkermansia. Initial experiments focused on chemical mutagenesis and later evolved into using a transposon system created by Andrew Goodman for use in Bacteroides11. Through a long process of trial and error, our first hint of success came from a mutagenesis experiment that generated just one mutant colony. Encouraged by this lone mutant, we continued to optimize our methods, eventually building the mutant library used in our study. The lab is continuing to develop genetic tools for Akkermansia and working towards a better understanding of this fascinating microbe.
References
- Derrien, M., Vaughan, E. E., Plugge, C. M. & de Vos, W. M. Y. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54, 1469–1476.
- Everard, A. et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. 110, 9066–9071 (2013).
- Derosa, L. et al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat. Med. 28, 315–324 (2022).
- Blacher, E. et al. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature 572, 474–480 (2019).
- Olson, C. A. et al. The Gut Microbiota Mediates the Anti-Seizure Effects of the Ketogenic Diet. Cell 173, 1728-1741.e13 (2018).
- Xie, J. et al. Akkermansia muciniphila protects mice against an emerging tick-borne viral pathogen. Nat. Microbiol. 8, 91–106 (2023).
- Davey, L. E. et al. A genetic system for Akkermansia muciniphila reveals a role for mucin foraging in gut colonization and host sterol biosynthesis gene expression. Nat. Microbiol. (2023) doi:10.1038/s41564-023-01407-w.
- Jia, B., Zou, Y., Han, X., Bae, J.W. & Jeon, C. O. Gut microbiome mediated mechanisms for reducing cholesterol levels: implications for ameliorating cardiovascular disease. Trends Microbiol. 31, 76–91 (2023).
- Bae, S., Mueller, O., Wong, S., Rawls, J. F. & Valdivia, R. H. Genomic sequencing-based mutational enrichment analysis identifies motility genes in a genetically intractable gut microbe. Proc. Natl. Acad. Sci. U. S. A. 113, 14127–14132 (2016).
- Nguyen, B. D. & Valdivia, R. H. Virulence determinants in the obligate intracellular pathogen Chlamydia trachomatis revealed by forward genetic approaches. Proc. Natl. Acad. Sci. 109, 1263–1268 (2012).
- Goodman, A. L. et al. Identifying Genetic Determinants Needed to Establish a Human Gut Symbiont in Its Habitat. Cell Host Microbe 6, 279–289 (2009).
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
Great work . Thanks for sharing.