Genetically engineered organoid models for the establishment of coronavirus drug targets
Published in Bioengineering & Biotechnology

March 2020. COVID-19 reached The Netherlands and the entire Hubrecht Institute went into lockdown, forcing all of us to work from home. Our lab, the Clevers group, has focused in the past decade on developing organoid models for many epithelial tissues. These organoids are adult stem cell (ASC) - derived 3D-structures grown in an extracellular matrix-mimicking gel and recapitulate key features of the corresponding organs. The versatile cultures have proven useful for unlocking secrets about development of epithelia, exceedingly rare cell types, diagnostic purposes and predicting therapy responses in patients. Adding layers of complexity to these cultures allows a reductionist approach to understand interactions between an epithelium and its surroundings. For example, immune cells can be added to tumor organoids to study efficacy of immune responses and therapy1. Pathogens can be injected into organoids to model the effect of individual species on human cells. This has for example contributed to our understanding that individual bacterial species can cause genetic alterations responsible for the development of colorectal cancer2.
After the first week of lockdown, when console games were getting less interesting, we started thinking how we could make any contribution to COVID19-research using our model systems. Meanwhile this could give us good excuses to return to the lab to do experiments. Early reports in February 2020 suggested COVID-19 sometimes causes severe gastrointestinal symptoms, particularly in children. Uncertain was if these effects were direct, caused by local infection, and if this would have any implications for viral spreading. Proteins that are essential for SARS-CoV-2 entry, including the cell surface ACE2, appeared to be expressed in specific cell types in the gut. In previous work, we had identified optimal cell culture conditions to produce such cells in organoids. Based on the expression of these proteins, the cells would be ideal candidates for presumptive infection by the virus.
We teamed up with the lab of Bart Haagmans in the Virology department of the Erasmus Medical Center in Rotterdam on March 14th. His lab has made key contributions to coronavirus biology in the past years, for example unraveling the key entry receptor for MERS-CoV. We set on a collaborative project in which we employed human intestinal organoids to assess cellular tropism of SARS-CoV-2, and measure the transcriptomic response to the virus. Multiple weeks later without any time off - a sharp contrast to the start of lockdown - and we had successfully performed SARS-CoV-2 infection experiments in organoids from different parts of the human gut. We mapped the immune response in these cells and found that enterocytes were very efficiently infected by the virus. This display of SARS-CoV-2 replication in human intestinal cells was published in May 2020 in Science, only 1.5 months after initiation of the project3. Two other studies published in the same month performed similar experiments to evidence the virus efficiently replicated in intestinal epithelial cells4,5.
After having established the intestinal tropism of SARS-CoV-2, we intended to use intestinal organoids to study more basic biology about viral entry and host factors. One of the main challenges in virology involves finding an appropriate in vitro model, as many viruses display restricted tropism. SARS-CoV-2 would for example not infect many of the classical animal models. Traditionally, most in vitro work depended on a kidney cell line derived from the African Green Monkey, Vero cells. This workhorse model has many advantages, including efficient infection (in fact, it is used to expand many viruses) and amenability to genetic engineering. However, this cell line does not fully recapitulate the biology of the affected cell types in humans. For example, chloroquine was found to be a good drug in Vero cells, but did not reach clinical efficacy in patients. Organoid cultures could potentially fill this gap, since many studies have shown their near-identical expression profiles to human epithelial cells. The most important sites of epithelial entry, airway epithelia and the intestine, can all be expanded as ASC-derived organoid cultures.
One key question is what factors viruses abuse to enter human cells, and replicate there. Any of such proteins could be appealing drug targets to influence viral spreading. CRISPR-Cas9-mediated gene editing had been applied in cell lines to pinpoint such factors. Although these cell lines allow for high-throughput approaches, translatability might be reduced due to the transformed nature of these cells. We opted to use intestinal organoids to build a biobank with mutants for previously identified host factors. Although a reduced number of genes could be assessed in these experiments due to technical limitations (transfecting organoids and growing clonal lines is time consuming), these are expected to better model the role of these factors in real epithelia.
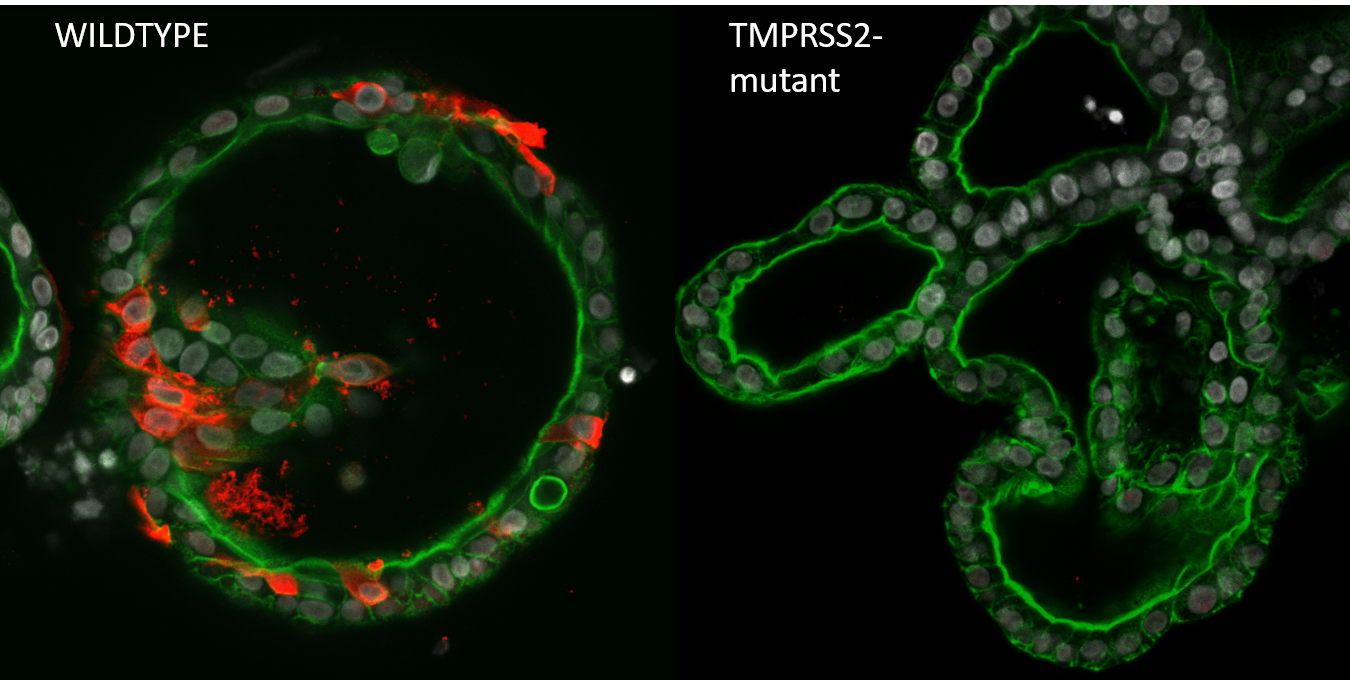
Starting in the summer of 2020, we started generating mutant organoids for previously published host factors. When mutants were generated, we would drive to the Erasmus MC where Mart Lamers (from the Haagmans Lab) would perform the Biosafety Level 3 part of the viral infection experiments. After dozens of car drives between Utrecht and Rotterdam, often on nearly empty highways in lockdown times, we had screened thousands of organoid clones for genetic alterations, and infected hundreds of mutant clones. Of the twenty genes that we had screened (the largest organoid knockout biobank to date), only a handful gave clear phenotypes on viral replication. While we could validate ACE2 to be the entry receptor and TMPRSS2 spike activating/priming protease of SARS-CoV-2 (see Figure), knocking out other host factors that were identified in high-throughput genome wide CRISPR screens (for example, NDST1 and NRP1) in organoids did not show a reduction in infection rate. Knocking out other TMPRSS family members (TMPRSS3, TMPRSS4, TMPRSS11D and TMPRSS13) did not affect virus replication, even though these genes were shown to activate the spike protein when over expressed in 293T cells6, indicating that there is remarkably little redundancy in the SARS-CoV-2 priming step. This may indicate that very specific TMPRSS2 inhibitors can be developed as therapeutic intervention.
We were also interested to see if we could validate the effectivity of proposed COVID-19 drug Chloroquine. Two putative entry pathways have been described for SARS-CoV-2: one utilizing cell surface TMPRSS2 to cleave (and activate) the spike protein, and the other involving receptor-mediated endocytosis followed by cleavage by cathepsins (including CTSL and CTSB). In our infection assays we did not see a reduction of SARS-CoV-2 infection rate upon knock-out of both cathepsins nor with the addition of chloroquine that inhibits this very pathway. Once again highlighting the use of organoids for coronavirus research. Our work was published in Nature Communications on September 17th.
Our experiments - and that of many others around the globe7–10 - have shown the utility of organoids as an attractive model to circumvent the issue of restricted tropism of many viruses. The progress in genetic engineering of organoids allows for identifying the role of host factors in cells closely resembling human epithelia, the viral point of entry. Our results differ from findings done in multiple cell lines. For example, TMPRSS2 is not mediating viral entry into Vero cells11, while specific TMPRSS2 inhibitors do appear effective in an animal infection model12. By generating this biobank we have the ability to quickly assess essential host factors for current and future coronavirus variants.
References
- Dijkstra, K. K. et al. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 174, 1586–1598.e12 (2018).
- Pleguezuelos-Manzano, C. et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature (2020). doi:10.1038/s41586-020-2080-8
- Lamers, M. M. et al. SARS-CoV-2 productively infects human gut enterocytes. Science (2020). doi:10.1126/science.abc1669
- Zang, R. et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. (2020). doi:10.1126/sciimmunol.abc3582
- Zhou, J. et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. (2020). doi:10.1038/s41591-020-0912-6
- Hoffmann, M. et al. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine (2021). doi:10.1016/j.ebiom.2021.103255
- Salahudeen, A. A. et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature 588, 670–675 (2020).
- Han, Y. et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589, 270–275 (2021).
- Youk, J. et al. Three-Dimensional Human Alveolar Stem Cell Culture Models Reveal Infection Response to SARS-CoV-2. Cell Stem Cell 27, 905–919.e10 (2020).
- Wagar, L. E. et al. Modeling human adaptive immune responses with tonsil organoids. Nat. Med. 27, 125–135 (2021).
- Wei, J. et al. Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection. Cell (2020). doi:10.1016/j.cell.2020.10.028
- Shapira, T. et al. A novel highly potent inhibitor of TMPRSS2-like proteases blocks SARS-CoV-2 variants of concern and is broadly protective against infection and mortality in mice. bioRxiv Prepr. Serv. Biol. (2021). doi:10.1101/2021.05.03.442520
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in