Getting a grip: The molecular mechanisms of Bdellovibrio attachment
Published in Microbiology, Protocols & Methods, and Cell & Molecular Biology

If Ridley Scott made a bacterium, he would probably create Bdellovibrio bacteriovorus. Its incredible lifestyle begins with a free swimming stage - Bdellovibrio is a small comma-shaped Gram-negative bacterium propelled by rotating a single flagellum. When Bdellovibrio encounters another Gram-negative bacterium (that is able to be preyed upon), it sequentially uses surface attachment, periplasmic invasion and sealing of the invasion “porthole”. It then releases a multitude of lytic enzymes that breakdown the prey bacterium from within, liberating biomolecules that Bdellovibrio consumes for filamentous growth. The elongated Bdellovibrio then septates forming multiple progeny, and lyses the prey to begin this cycle again.1,2
Fig 1 - The Lifestyle of Bdellovibrio bacteriovorus. Free swimming Bdellovibrio attach to prey and invade into the periplasmic space. The prey cell rounds up forming the bdelloplast and Bdellovibrio consumes the prey to drive filamentous growth. The predator then septates and lyses the bdelloplast to release progeny.
Bdellovibrio is a truly incredible organism to study, as it has a wealth of intriguing machinery that it needs to enable this novel lifestyle. The predatory lifestyle also makes Bdellovibrio and like organisms (BALOs) effective candidates for biotechnology and biological control of bacteria in health and industrial settings.3,4 Having a fundamental understanding of the molecular mechanisms underpinning this lifestyle is crucial for its effective use as a 'biological antibiotic' and could lead to novel therapeutics and diagnostics derived from the molecular machinery involved.
Although B. bacteriovorus’ identification had its 60th anniversary in 2022, there are still huge gaps in our understanding of the mechanisms involved in predation. In particular, there has been no identification of any molecular entity involved in the attachment of the predator to its prey. Through an effective collaboration between the Lovering group at the University of Birmingham and the Sockett group at the University of Nottingham we have used a combination of complementary techniques to crack a hole into the elusive nature of predator-prey recognition. In our paper, we identify and characterise a novel family of adhesins from B. bacteriovorus, which we termed mosaic adhesive trimeric (MAT) fibres.
The inception of this project was elicited by a serendipitous finding by the Sockett group at the University of Nottingham, where they were fluorescently tracking Bd0635, a CpoB homologue in Bdellovibrio presumed to be involved in cell wall biosynthesis. They observed that Bdellovibrio 'pinches off' a portion of itself to produce a vesicle-like structure within invaded cells. Vesicles could be isolated and purified, and proteomic studies showed that they contained an abundance of invasion-associated proteins. In this list were a subset of the 21-strong MAT family, which are peppered throughout the genome and the full group was later identified through sequence homology to the initial proteomics hits.
The Sockett group subsequently investigated the localisation of these fibres and proved their role in predation using in vivo fluorescence microscopy. Knocking out the genes of some of these fibres causes a small defect in predation, although it’s difficult to observe strong effects when there is such a high redundancy of MAT fibres. Interestingly, MAT location was shown to vary – present during attack of prey (at the biting pole) but also at the side wall and flagellar pole. After invasion MAT localisation is altered and some of these fibres were sequestered to the vesicle, indicative of usage at different points of contact during predation.
The Lovering group undertook the biochemical and structural characterisation of the MAT proteins, aiming to probe the features behind specificity; it was intuitive that fibre specificity would occur at the distal “tips”. The majority of MAT fibres terminate in a C-terminal S74 protease domain (typically comprising ~10% of the fibre length), which is a feature commonly found in phage tail fibres and acts as an autoproteolytic chaperone that enables folding of the interdigitated trimers. This domain cleaves itself from the fibre tip and leaves a mature protein with a C-terminal beta-helix - three elongated sheets that spiral round the central axis of the fibre. Unlike many other trimeric fibres such as coiled coils, these are beta-rich and produce a highly stable structure. The first structure we solved was that of the S74-containing MAT protein Bd3182 (Fig 2), which we believe will bind to prey surface molecules using a cleft formed by the beta sheets and loops, similar to that of phage interactions with their prey.
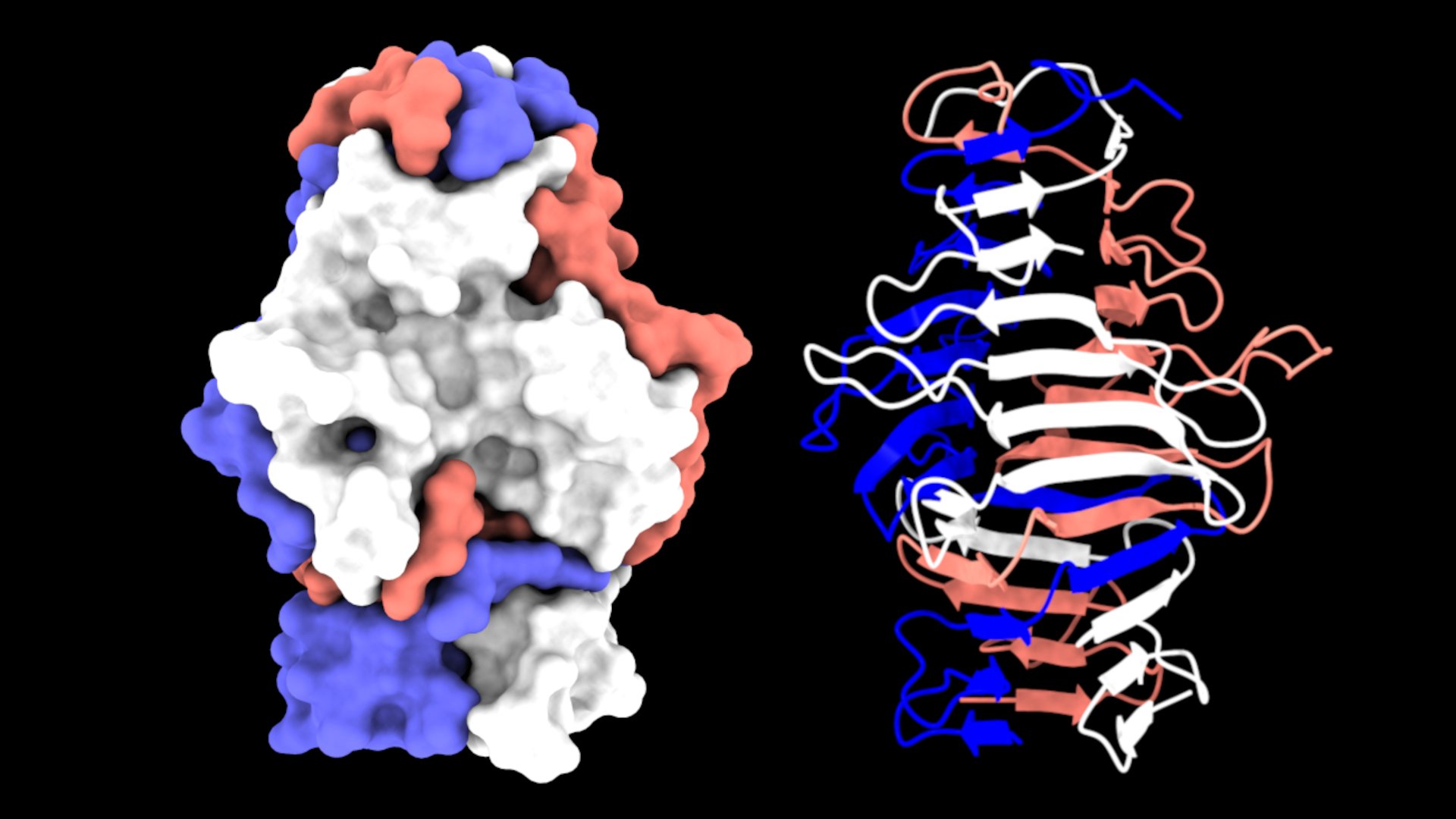
Fig 2 - The structure of the MAT Bd3182 tip. The C-terminal region of the mature Bd3182 fibre displays the classical twisting trimeric beta-helix. Each of the three monomers that form the fibre are shown in different colours.
Interestingly, Bdellovibrio bacteriovorus strain HD100 has 8 MAT proteins that switch the S74 domain for an alternative fold at the terminus. To investigate why B. bacteriovorus uses different termini, the Lovering group solved the structure of five C-terminal regions of these representatives. The alternate domains are hugely diverse and include tumour necrosis factor (TNF)-like domains, lectin domains, and multiple novel folds of unknown function. These latter proteins may represent adhesive domains that have yet to be characterised in biology. In most cases these tips are preceded by a similar beta-helix domain to that of the S74-containing MAT fibres. This mosaic swapping of domains is likely to ensure that Bdellovibrio can adhere to a wide range of molecular targets on the surface of diverse prey bacteria. In addition to our experimental data, we used modelling techniques to predict the structure of the full-length MAT fibres and show that in addition to the C-terminal domain, they contain a common N-terminal domain, and elongated interspersing regions between the two (Fig 3). Hence MAT fibres have a common (anchoring) region, a more diverse central linker, and a very diverse use of domains at the distal prey-recognition end – intuitive for a bacterium that recognises a multiplicity of prey.
We also managed to elucidate the molecular targets of Bd2439, one of the TNF-terminating non-S74 MATs. We subjected the C-terminus of Bd2439 to a glycan array and observed that this protein adhered to LPS from 6 diverse strains of bacteria, but particularly strongly to that of Proteus mirabilis. This observation was very surprising because the HD100 strain of B. bacteriovorus was originally isolated from soil and would never usually come into contact with these LPS. The fibre redundancy, multiple LPS specificity, and the inherent ability to bind molecules from non-native environments highlights that Bdellovibrio and its MAT fibres are evolutionary optimised for versatility.

Fig 3 – Select MAT Fibres of B. bacteriovorus; 6 modelled fibres are shown (each containing a C-terminal region that we experimentally solved and verified). The similar N-termini can be seen on the left side, and the diverse tip domains can be seen on the right side.
MAT fibres are not specific to B. bacteriovorus and can be found in related predatory bacteria such as other BALOs and the related myxobacteria. Amazingly, they can also be found in completely unrelated predatory and scavenging bacteria like the prey-cell-engulfing Candidatus Uabimicrobium amorphum and the cyanobacteria-eating Salinivirga cyanobacteriivorans. These findings emphasise the role of MATs in diverse lifestyles.
In summary, we have uncovered the means by which Bdellovibrio predators have adapted to recognise a multiplicity of prey with diverse surfaces, and provide a toolkit from which the community can now investigate individual specificities. Perhaps one day we will be able to engineer predatory bacteria with designed killing ranges, or at least ones that show a bias towards those we wish to remove.
References
1. Lovering AL, Sockett RE. Microbe Profile: Bdellovibrio bacteriovorus: a specialized bacterial predator of bacteria. Microbiology (Reading). 2021 Apr;167(4):001043. doi: 10.1099/mic.0.001043. PMID: 33843574; PMCID: PMC8289219.
2. Sockett RE. Predatory lifestyle of Bdellovibrio bacteriovorus. Annu Rev Microbiol. 2009;63:523-39. doi: 10.1146/annurev.micro.091208.073346. PMID: 19575566.
3. Bratanis E, Andersson T, Lood R, Bukowska-Faniband E. Biotechnological Potential of Bdellovibrio and Like Organisms and Their Secreted Enzymes. Front Microbiol. 2020 Apr 15;11:662. doi: 10.3389/fmicb.2020.00662. PMID: 32351487; PMCID: PMC7174725.
4. Cavallo FM, Jordana L, Friedrich AW, Glasner C, van Dijl JM. Bdellovibrio bacteriovorus: a potential 'living antibiotic' to control bacterial pathogens. Crit Rev Microbiol. 2021 Sep;47(5):630-646. doi: 10.1080/1040841X.2021.1908956. Epub 2021 May 1. PMID: 33934682.
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in