GO2Sum: Generating Human Readable Functional Summary of Proteins from GO Terms
Published in Protocols & Methods
Proteins are the building blocks of life, fulfilling crucial roles within living organisms such as signal transduction, catalyzing metabolic reactions, and maintaining cellular structure. Determining the function of a protein is one of the key challenges in modern biology and bioinformatics given its plethora of downstream applications involving drug discovery and the understanding of disease and organic mechanisms. Traditionally, Protein function prediction (PFP) involves predicting a set of functions a protein is likely to perform, which can comprise a large list, sometimes even up to 50-100 functional terms (Fig. 1a). To represent protein functions, Gene Ontology (GO), a controlled vocabulary, is frequently used because it is easy to handle by computer programs, avoiding open-ended text interpretation. GO includes three sub-ontologies for describing molecular functions (MF) of a single protein, biological processes (BP) to which proteins can contribute, and cellular components (CC) where proteins are active.
.png)
Figure 1.Background for GO2Sum a, protein function prediction pipeline b, The workflow for the retrieval of Function, Subunit Structure, and Pathway paragraphs from UniProt.
Therefore, we approached the function prediction problem from a different perspective, aiming to make the functional descriptions human readable. We utilized a large language model (LLM), T5, which was pre-trained on the entire web corpus Colossal Cleaned Common Crawl (C4) dataset, thus possessing an understanding of both human language and biological information. We then fine-tuned the model so that it could generate three different functional summaries: "Function" (general function description, usually presented at the top of the UniProt entry), "Subunit Structure" (detailing molecular interactions with other proteins), and "Pathway Information" paragraphs (explaining the metabolic pathways associated with the protein) when given all the functional descriptions as input. Fig. 1b shows examples of these three function summaries from the UniProt database for protein A9AJN2 (Phosphatidylserine decarboxylase proenzyme).
Our generated functional summaries are highly readable and closely resemble the UniProt functional summaries provided by experts
Fig. 2, illustrates the workflow of GO2Sum, It begins with a set of input GO terms, each accompanied by a text description. For instance, GO:0000049 is associated with the description ‘Binding to a transfer RNA.’ These GO term descriptions are concatenated into a document, serving as the input for the summarizer model, T5. The summarizer then generates a paragraph that elucidates the function of the input protein.
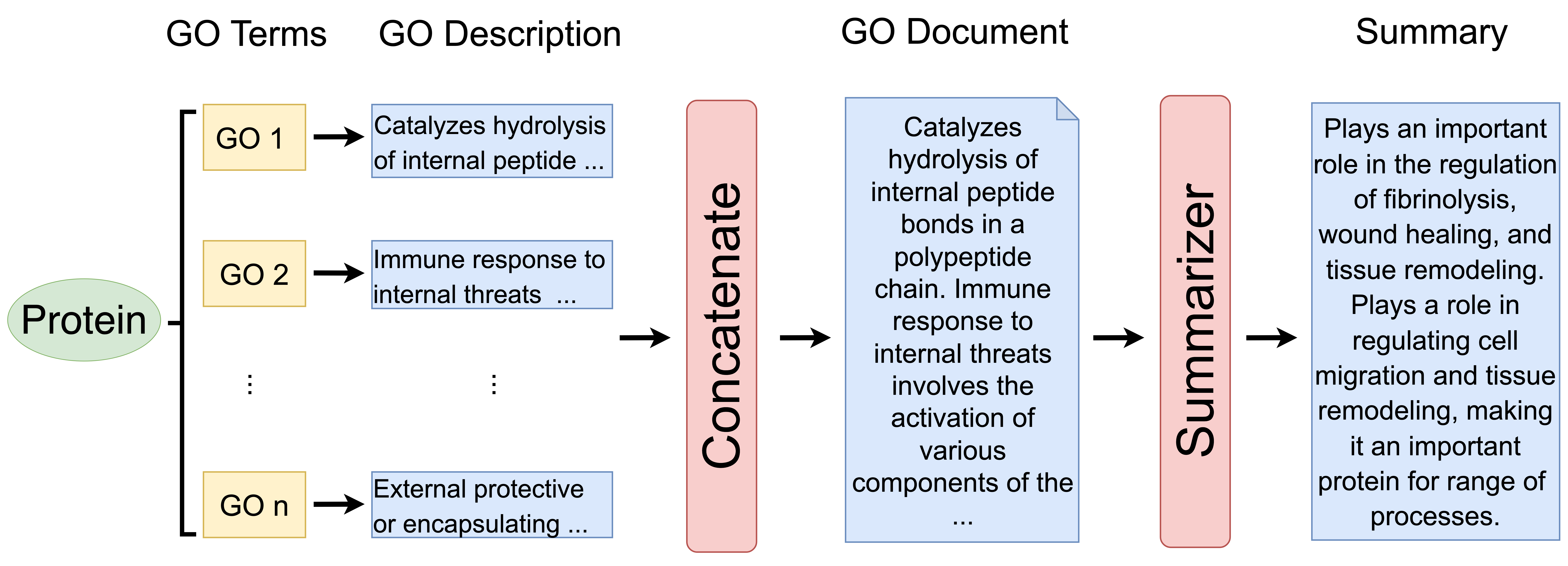
Figure 2: GO2Sum workflow
In Table 1, we provide several examples of generated summaries for UniProt Function. GO2Sum produced summaries that closely matched the UniProt paragraphs, with negligible differences. These differences were generally minor, such as the omission of specific protein names or slight variations in wording with the same meaning. In the first example, P0DMN7 ( S-adenosylmethionine decarboxylase proenzyme 1), the UniProt description has several keywords, polyamines, spermidine, spermine, and embryonic. GO2Sum was able to identify all of these keywords. In contrast, T5 produced incomplete phrases and only identified ‘spermine’ and ‘spermidine’.
Interestingly, there were also cases where GO2Sum output function description that is more accurate and informative than the UniProt function description, protein P60154 (Inactive ribonuclease-like protein 9) is such an example UniProt describes its function only as 'Does not exhibit any ribonuclease activity,' which is not a comprehensive summary of the known functional activity of this protein. This protein has a GO term annotation, GO:0060474, which is 'The process in which the controlled movement of a flagellated sperm cell is initiated as part of the process required for flagellated sperm to reach fertilization competence', which is more informative. This is captured by GO2Sum and produced the summary around it. GO2Sum captured the main concept of the protein’s function, which is the involvement in sperm motility and acts as a signaling molecule necessary for the proper movement of spermatozoa.
Table 1: Examples generated by GO2Sum and Vanilla T5 for UniProt Function
| Protein Name | ID | UniProt | T5 | GO2Sum |
| adenosylmethionine decarboxylase proenzyme 1 | P0DMN7 | Essential for the biosynthesis of the polyamines spermidine and spermine. Promotes maintenance and self-renewal of embryonic stem cells, by maintaining spermine levels. | spermine. spermidine. Binding to putrescine | Essential for biosynthesis of the polyamines spermidine and spermine. Promotes maintenance and self-renewal of embryonic stem cells. |
| Peroxisome assembly protein 12 | Q9ET67 |
Required for protein import into peroxisomes. |
a zinc ion (Zn). a peroxisome is |
Somewhat implicated in the biogenesis of peroxisomes. |
| Inactive ribonuclease-like protein 9 | P60154 | Does not exhibit any ribonuclease activity | a nucleic acid. space outside of the plasma membrane. For cells without |
Involved in sperm motility. Seems to act as a signaling molecule which is required for the flagellated sperm to enter fertilization competently. |
Through this work, we are embarking on a new direction in bioinformatics research, leveraging the recent advancements in LLM capabilities to translate these GO term predictions into human-readable text, providing a more intuitive and user-friendly approach to describing predicted functions. GO2Sum may be useful for various tasks that deal with GO terms and functions, such as automated generations of function descriptions for newly annotated genomes, function predictions, and identification of GO terms from text from literature.
To contribute to GO2Sum, we have released our source codes at https://github.com/kiharalab/GO2Sum . If you have any questions or potential ideas to further improve GO2Sum, please contact Daisuke Kihara (dkihara@purdue.edu).
Follow the Topic
-
npj Systems Biology and Applications

An online Open Access journal dedicated to publishing the premier research that takes a systems-oriented approach and encourages studies that integrate, or aid the integration of, data, analyses and insight from molecules to organisms and broader systems.
Related Collections
With Collections, you can get published faster and increase your visibility.
Next-Generation Mammalian Cell Bioprocessing: Systems Biology, Synthetic Biology, and Beyond
Publishing Model: Open Access
Deadline: Mar 15, 2026
Systems mechanobiology
Publishing Model: Open Access
Deadline: Mar 28, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in