Graph Fourier transform for spatial omics representation and analyses of complex organs
Published in Protocols & Methods and Statistics

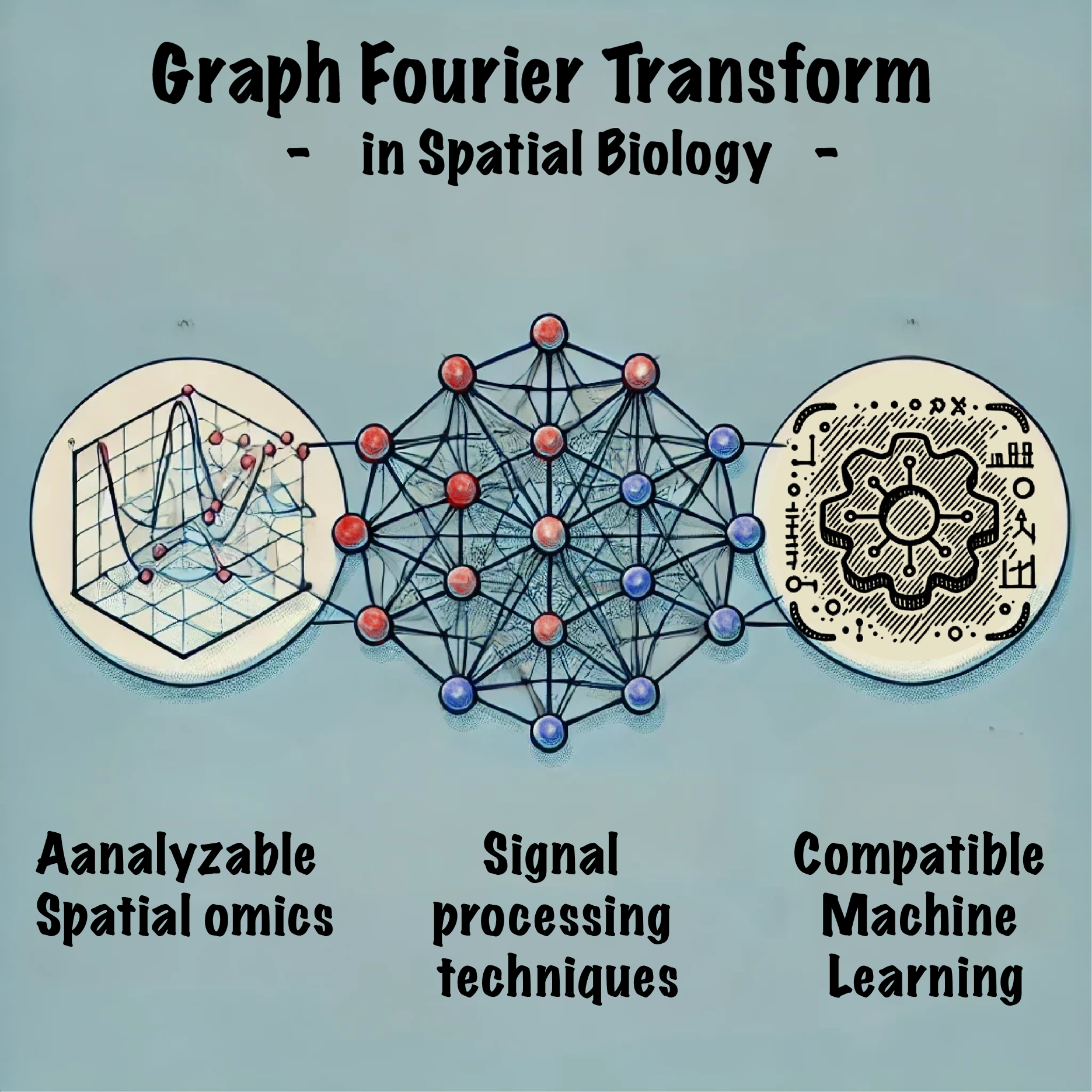
Concept of graph Fourier transform
Graph Fourier Transform (GFT) is an advanced analytical method inspired by the traditional Fourier Transform, adapted to analyze data within the structure of graphs. This technique provides a quantitative way to represent signals on a graph, enabling a thorough analysis of complex data sets. In GFT, data points (nodes) that represent cells or spots and their interconnections (edges defined by spatial coordinates) are analyzed to break down complex biological signals. This transformation translates graph signals (e.g., spatial omics features) into spectra that uncover intrinsic patterns and variations, facilitating several applications,
- Analyze the properties of graph signals.
- Filter noise and enhancement of important feature data signals.
- Connect to machine learning and deep learning
The importance of GFT in spatial omics
GFT is essential in spatial omics for its ability to transform complex omics features into another feature space that is not only interpretable but also analyzable. By organizing data into low, medium, and high-frequency components, GFT facilitates an understanding of biological data: low-frequency components reveal overarching biological trends, medium-frequency components capture transitional features within tissues, and high-frequency components pinpoint localized phenomena. Each bandwidth thus serves a distinct and valuable role in dissecting and understanding tissue samples more comprehensively.
Exploring and reporting the GFT in our work
We introduced Spatial Graph Fourier Transform (SpaGFT), a new analytical method that enables the representation and analysis of spatially organized biological data. We demonstrated the following applications of SpaGFT on spatial omics data.
- Our approach formulates the identification of spatially variable genes (SVGs) as a k-bandlimited signal recognition problem, and performance outperformed other tools.
- We employ a low-pass filter and inverse GFT to enhance the signal by amplifying the low-frequency components while suppressing noise, leading to enhancing spatial domain identification.
- Transformed features by SpaGFT can be clustered and derive spatial domain, which is allowed to be overlapped, revealing more complex spatial domains that are not limited by rigid boundaries.
- We implement SpaGFT to high-resolution CODEX data and reveal morphological and molecular diversity in secondary follicles, such as mantle zones and germinal centers, showcasing B and T cell organization and interactions.
- SpaGFT can be integrated into existing machine learning frameworks to enhance their performance. We have implemented this approach in four spatial tools: SpaGCN, Tangram, TACCO, and CAMPA, demonstrating substantial improvements.
- SpaGFT provides bandwidth guidance to CAMPA. The model gains the ability to identify rare subcellular structures, such as the Cajal body and Set1/COMPASS complex, highlighting the potential of our method to refine and advance spatial omics analysis.
Future perspectives
Looking ahead, the future development of SpaGFT could focus on several key areas to enhance its functionality and expand its impact in spatial omics research. First, exploring medium- and high-frequency signals could uncover additional layers of biological complexity by capturing localized phenomena and rapid responses to environmental stimuli. Enhancing computational efficiency would enable faster analysis of larger datasets. Addressing the challenges of cross-sample comparability, future efforts may develop methods to align spatial samples into a common reference space, potentially using machine learning frameworks or histology images. Automating the detection of functional regions in multiplexed images through topological learning frameworks could further streamline the analysis process. Additionally, integrating SpaGFT with explainable machine learning models could enhance their ability to identify complex biological structures and rare subcellular organelles by providing augmented features and regularizers. Ultimately, expanding SpaGFT's applications to multi-resolution spatial omics data integration and spatiotemporal pattern analysis could provide deeper insights into complex tissue biology.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
new publish