Harmonized Datasets of microbiological analyses from a French national-scale soil monitoring survey
Published in Earth & Environment, Ecology & Evolution, and Microbiology

Soils support many ecosystem services (eg fertility, carbon storage or waste decomposition). All these services are mainly supported by the huge reservoir of soil microbial biodiversity. However, soil microbiota is constantly subjected to various natural and/or anthropogenic stresses (eg deforestation, land-use intensification and global warming). These disturbances have a significant influence on these soil microbial communities and lead to an overall impact on soil functions. For the past 15 years, the French Soil Quality Monitoring Network (RMQS) has been meeting these goals of long-term assessment and monitoring of soil quality in France.
The RMQS is based on the monitoring of 2,240 sites distributed across the whole French territory along a systematic square grid of 16 km x 16 km cells, to be representative of the different types of soils and their land uses. Soil sampling, characterization and observations are made every 15 years at the center of each cell. The RMQS is probably one of the most intensive and extensive sampling strategy at a national scale in Europe.
Thanks to the use of various molecular tools, a substantial body of scientific knowledge has been produced on the RMQS soil microbiota.
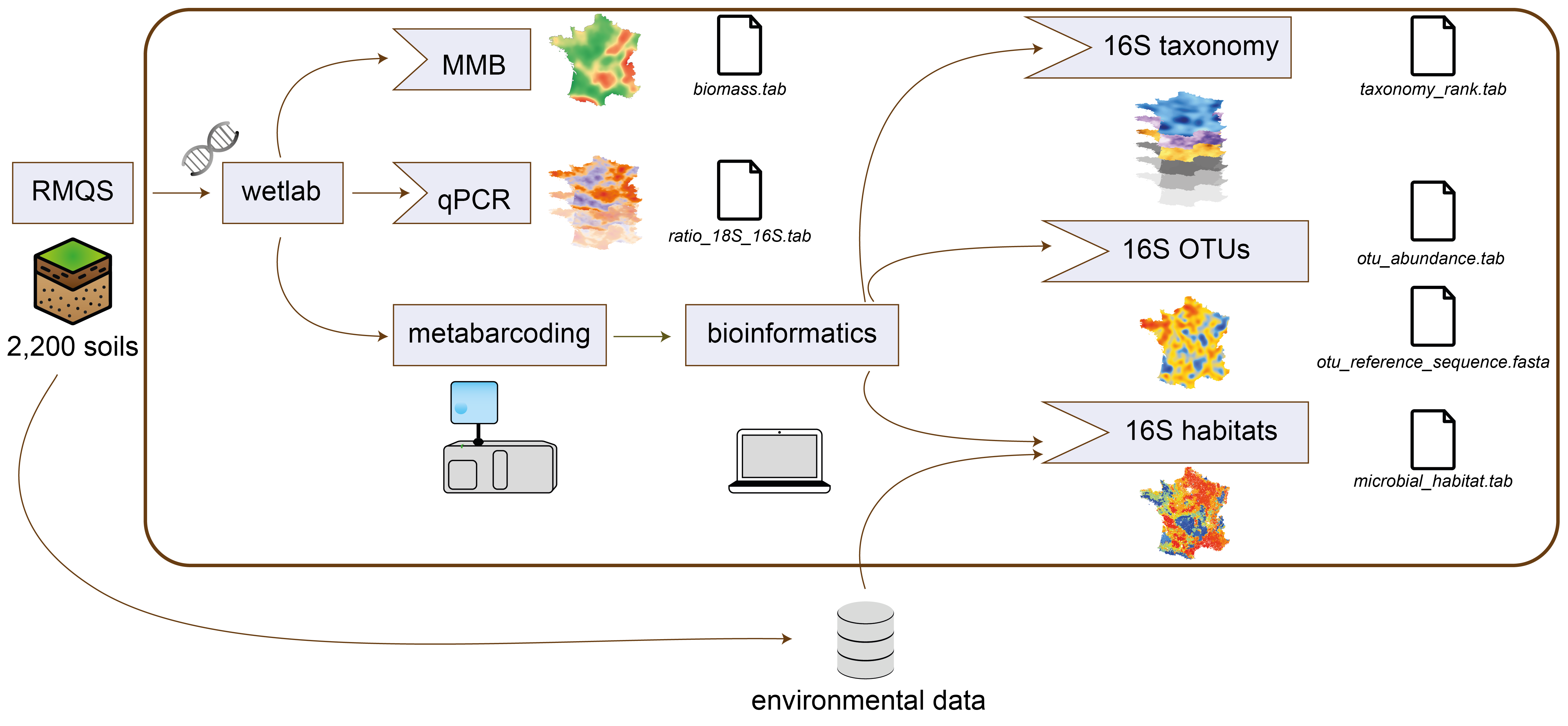
Schematic overview of the RMQS1 microbial datasets.
Interestingly, each produced dataset (e.g. measures of 16S and 18S gene abundances, the F:B ratio, or the diversity data) exhibited different and specific biogeographical patterns compared to each other taken separately, reflecting each one a complementary snapshot of soil microbial communities. To improve the reuse of these datasets, we harmonized and reorganized all available microbiological in a Dataverse collection, with a focus on linking datasets with other RMQS environmental parameters more easily.
This collection will provide information on the ecology of microbial communities at a territorial scale and describe the different microbial groups observed in French soils, their spatial distribution, their ecological requirements and their interactions. Altogether, this collection will help researchers to have a better understanding of soil microbial communities organization and dynamics across space at a national scale.
Follow the Topic
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
Publishing Model: Open Access
Deadline: Apr 17, 2026
Data to support drug discovery
Publishing Model: Open Access
Deadline: Apr 22, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in