In our busy lives, we often struggle with prioritizing tasks. Bacteria and archaea, it turns out, also face the same issue when thwarting attacks by invaders. These invaders, usually mobile plasmids or bacteriophages, sneak into host cells with the purpose of hijacking the cell’s machinery and resources to reproduce. Host cells in turn have developed efficient immune systems that recognize these invaders in hopes of blocking the infection and preventing further spread. As the only adaptive immune system in bacteria and archaea, CRISPR-Cas systems store genetic information of invaders as spacers in between repeated sequences on CRISPR arrays, where new spacers are sequentially added at one end of an array. The stored information is then recalled as CRISPR RNAs (crRNAs) guiding Cas nucleases to cleave the invader’s genetic material or enact widespread collateral cleavage of RNA to shut down the infected cell.
As one can imagine, the CRISPR arrays of some long-suffering bacteria/archaea can grow to include tens or even hundreds of spacers. The newly acquired spacers represent a weapon against recently encountered invaders which are likely still present in the environment, while older spacers likely match to invaders that have mutated the target to avoid detection or are no longer present in the environment. Large CRISPR arrays pose a problem though: producing large numbers of crRNAs creates competition for the limited processing machinery and Cas nucleases. The bigger threat of recently encountered invaders and potential competition for the Cas machinery underscore the importance of prioritizing the newest spacers for immune defense. Prioritizing spacers is also compatible with the well-known life motto of microbes – being energy efficient is the key to surviving in hard times. Indeed, people have noticed that new spacers always give rise to more abundant crRNA. However, the underlying mechanism has remained elusive. As a postdoc researcher who worked in Dr. Chase Beisel’s lab in Helmholtz Institute for RNA-based Infection Research, I uncovered the mystery with help of colleagues.
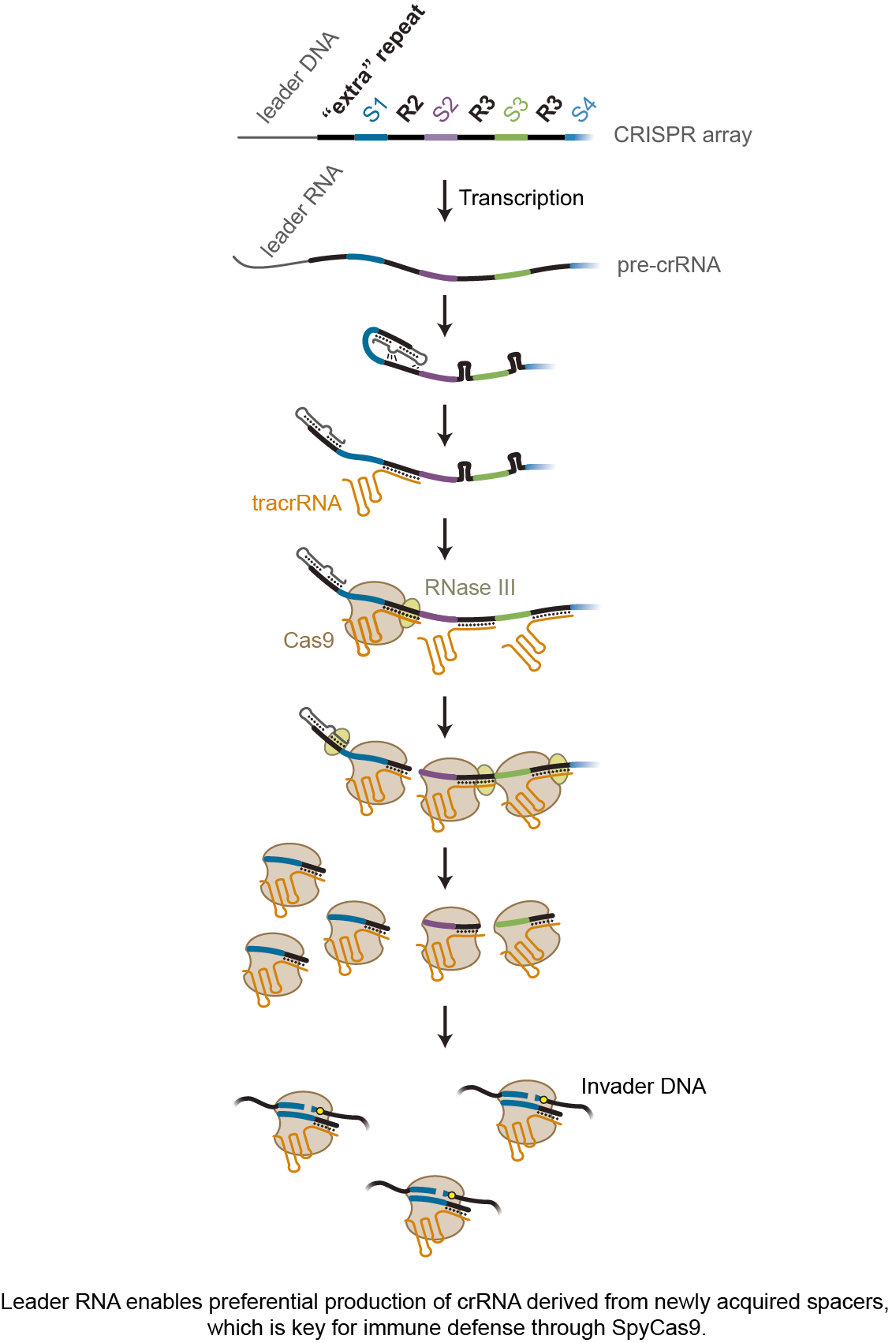
Our study started with our curiosity about the fate of the first repeat on the endogenous CRISPR array found in Streptococcus pyogenes, the source of SpyCas9 widely used for genome editing applications. This “extra” repeat is supposed to give rise to a crRNA, only the targeting sequence is derived not from a spacer but the upstream sequence of the array called the leader. We found this unintended crRNA was not produced. While looking for possible reasons, we found that the transcribed leader-repeat was forming a stable stem-loop which inhibited the production of the unintended crRNA. Intriguingly, mutating the leader to open the stem-loop structure disabled anti-phage and anti-plasmid activity mediated by the first spacer on the array. We then reached out to Dr. Cynthia Sharma from the University of Würzburg and just down the hall, who helped conduct RNA sequencing of crRNAs bound to Cas9. The result confirmed that mutating the leader significantly reduces the crRNA abundance of the first spacer. Meanwhile, an RNA species spanning the leader and the second repeat caught our attention. As part of crRNA biogenesis, the repeats in the transcribed array base pair with another RNA encoded near the CRISPR array – tracrRNA. The RNA duplex is then cleaved by RNase III and bound by Cas9, which gives rise to individual crRNA. The accumulated RNA spanning the leader through the second repeat suggested that the second repeat on the array is preferentially processed. We also mutated the leader in different ways, finding that the loop regions on the original leader-repeat stem-loop was the key in the preferential processing of the second repeat.
Therefore, we hypothesized that the loops interact with the second repeat, which promotes base pairing between the second repeat and the tracrRNA to yield accelerated processing of this repeat and formation of the crRNA. To test this hypothesis, we reached out to Dr. Neva Caliskan in our same institute and also located down the hall, who helped conduct Microscale thermophoresis. These experiments confirmed that mutating the loop reduces binding of the tracrRNA with the second repeat. As a final test, we mutated the loops and the second repeat to disrupt and restore these interactions, which disabled and then restored anti-plasmid activity through the first spacer. These results supported the leader interacting with not only the first repeat but also the second repeat to accelerate processing of the crRNA derived from the newest spacer.
Seeing this remarkable instance of prioritization in S. pyogenes, we asked whether this phenomenon exists across different CRISPR-Cas systems. We reached out to Dr. Rolf Backofen at the University of Freiburg and Dr. Zasha Weinberg at Leipzig University, neither down the hall but still good collaborators within a train-ride, who helped extract leader-repeat sequences from public databases and predict the probability of stem-loop formation. They found that leader-repeat stem-loop appears prevalent across type II-A CRISPR-Cas systems, the subtype in which the system in S. pyogenes is found. However, this leader-repeat stem-loop was virtually absent elsewhere. Besides SpyCas9, we also investigated and confirmed the formation of leader-repeat stem-loops in native CRISPR arrays from II-A CRISPR-Cas systems in Lactobacillus rhamnosus and Streptococcus thermophilus.
To conclude, we believe our study is the first to reveal how CRISPR-Cas systems prioritize the most recently acquired spacers for immune defense. Our hope is that our work spurs others to investigate alternative mechanisms deployed to prioritize new spacers across the diverse CRISPR-Cas systems. Our study also revealed a new role of the leader RNA in crRNA biogenesis, which might be further extended beyond II-A CRISPR-Cas9 systems to other systems as hinted by our bioinformatic analyses. Another intriguing element we discovered is that the interaction between leader-repeat stem-loop and downstream repeat promotes binding of tracrRNA to the downstream repeat. We collected solid data supporting the existence of these interactions and their contributions to immune defense by SpyCas9, although more efforts are needed to resolve the specific structure. Regardless of the details of the interactions, this simple but elegant mechanism is perfect for facilitating immune defense by SpyCas9. Besides improving our understanding of fundamentals of CRISPR biology, the knowledge gleaned from our study will also drive efforts to engineer CRISPR arrays for multiplexing applications of CRISPR technologies.
References:
Deltcheva, E. et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607 (2011).
Jackson, S. A. et al. CRISPR-Cas: Adapting to change. Science 356, (2017).
McGinn, J. & Marraffini, L. A. Molecular mechanisms of CRISPR-Cas spacer acquisition. Nat. Rev. Microbiol. 17, 7–12 (2019).
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in