
It is cold in the depths of the world’s oceans; most of the seafloor and overlaying water is a chilly 4 °C. However, this is not so at the seafloor of Guaymas Basin in the Gulf of California. There, the Baja California peninsula is separating from North America, and heat from Earth’s interior rises up. Right at the seafloor where the geothermal heat meets the cold deep ocean, the sediments often reach a cozy 60 °C, an ideal temperature for heat-loving microbes. It appears that many of these heat-loving microbes rely on methane, an abundant energy source transported upwards from deeper sediment layers by hydrothermal fluids. These methane munchers have received significant attention given their vital role in methane removal and carbon fixation in the deep-sea. However, the charismatic methane munchers make up only a small portion of the overall microbial diversity. Many other heat-loving microbes rely on larger organic compounds and – like humans – use sugars, proteins, and fatty acids as an energy and carbon source. We decided to put the spotlight on these heterotrophic heat-loving microbes and the organic macromolecules they like to devour.
.jpg)
Methane munchers at a hydrocarbon sediment tower seen from Alvin at ~2000 m depth in Guaymas Basin. Photo Teske et al. 2021
It took 4 countries, 5 women and 3 men scientists, and their collaborators to make this happen. It all started in Canada where Emil Ruff, a postdoc at the time with Marc Strous (University of Calgary), was invited by Andreas Teske (University of North Carolina) to submit a research idea in order to participate in the 2016 Guaymas Basin expedition.
“The idea of looking at biological macromolecule degraders originated from a global study in which we realized that the majority of microbial diversity at methane seeps worldwide belongs to heterotrophic lineages.” – Ruff
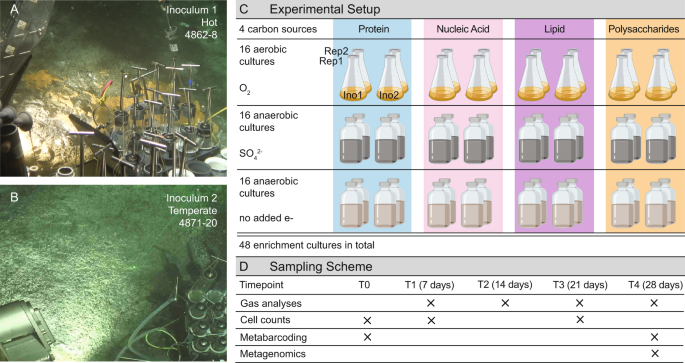
In 2018 Emil started a position at the Marine Biological Laboratory (Woods Hole, MA), where he brought Kathleen Regan and me on his team to take this work to the finish line. In addition to the gas chromatography and 16S rRNA analyses (assisted by Isabella) Emil performed on 48 enrichment cultures at the Strous Lab, he planned to incorporate cell counts (performed by Kathleen) and metagenomics. As a field-based soil ecologist, I needed training to process ~130,000,000 DNA sequencing reads from these cultures and decided to participate in the Workshop on Genomics, Cesky Krumlov. At the workshop, I met Kelly Wrighton who was an invited speaker and - over Aperol refreshers - told me that I could characterize the diets of the 20 genomes I assembled with a friendly and fast tool she and her collaborators created. Kelly and Mikayla Borton (Kelly's PhD student at the time) not only introduced me to DRAM but helped me to describe our heterotrophic heat-loving microbes.

Alvin’s robotic arm sampling heterotrophic heat-loving microbes by an orange Beggiatoa mat. Pérez Castro et al. 2021
We showed that certain heat lovers specialize on degrading the biological macromolecules that make up all living cells - from bacteria to humans. When cells perish and burst these macromolecules – also called necromass - are spilled to the environment and become available as a food source for other organisms.
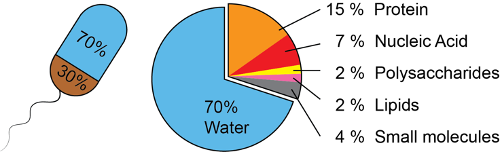
Every living cell is made up of roughly the same molecules and compounds. Four compound classes are responsible for most of the biomass: proteins, nucleic acids (mostly DNA and RNA), polysaccharides (e.g., starch, cellulose) and lipids.
At Guaymas Basin some of the necromass is supplied from the overlying water column, but also from the methane munchers and other primary producers at the base of the food web. We found that the macromolecule eating microbes in our cultures belonged to previously uncultivated microbial species and that these organisms also occurred at other hot ecosystems like the black smokers of the mid-Atlantic Ridge. The experiments also showed that each compound sustained a whole food web of microbes, which could explain the high diversity of coexisting heterotrophs. To our surprise, none of the cultures in the 24 different tested conditions produced methane, a common end product of anaerobic microbial communities. This could mean that the subsurface-derived greenhouse gas that is removed by the methane-munching microbes at the seafloor is not produced again by the heterotrophic anaerobes. Instead, it seems to be removed from this deep-sea ecosystem for good, which has important implications for our understanding of the deep-sea carbon cycle.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in