Hidden in the blood – can blood microbiomes flag potential disease outbreaks?
Published in Microbiology

African buffalo are reservoirs for a variety of diseases such as bovine tuberculosis, brucellosis, Rift Valley fever, tick-borne diseases and foot and mouth disease. Richard Nyamota and his colleagues studied the blood microbiome of clinically healthy buffalo to map out their ‘bacterial and pathogenic landscape’.
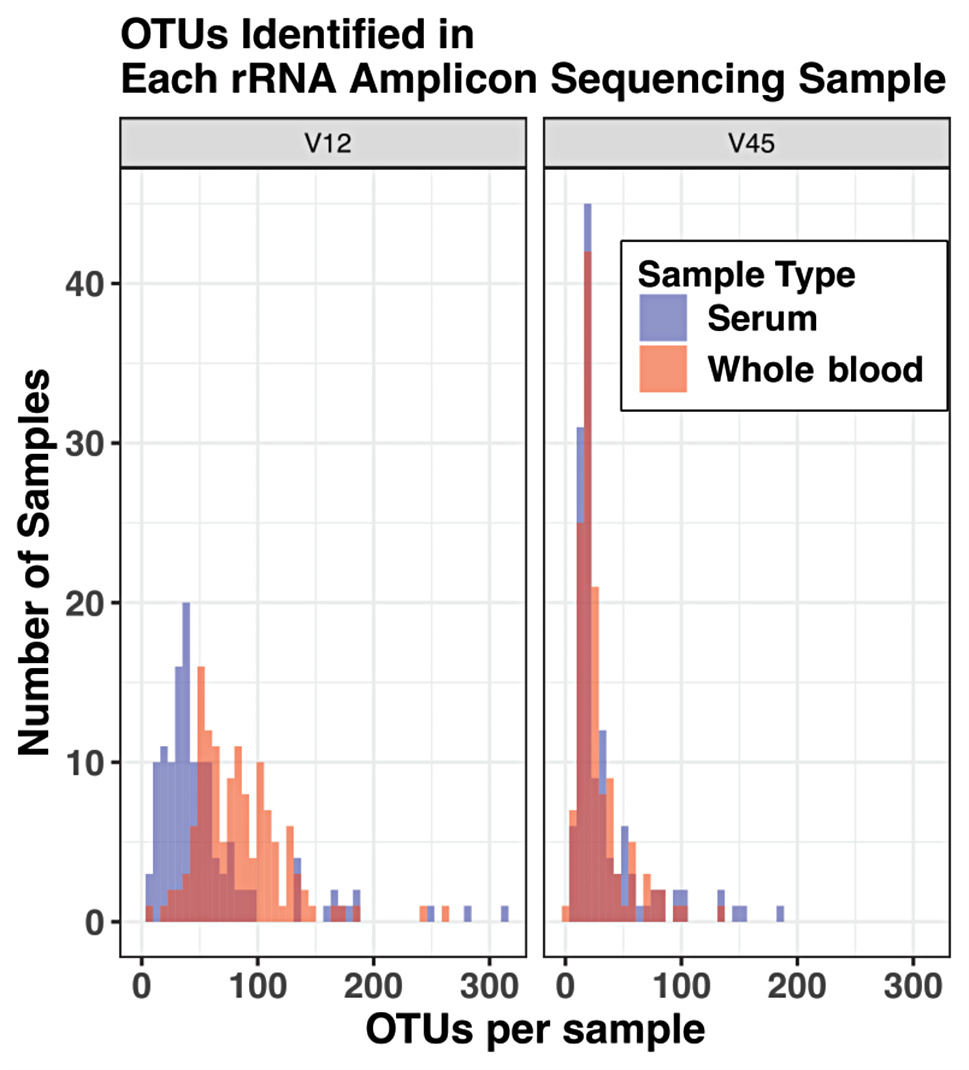
They collected whole blood and serum samples from 46 wild African buffaloes in Kenya, and sequenced the total DNA extracted from the samples to identify the bacteria present. They found a diverse community of bacterial species in both whole blood and serum samples, but Rickettsiales and Mycoplasmatales were the most abundant - Rickettsiales being more predominant in whole blood samples, whilst Mycoplasmatales were predominant in serum samples. Rickettsiales are an order of bacteria that are obligate intracellular parasites found in arthropods and mammals and are of One Health significance.
In their study, Richard Nyamota and colleagues found Anaplasma to be the most abundant of the Rickettsiales – the genus of bacteria causes anaplasmosis in humans and mammals (transmitted by tick bites) and can cause organ failure and death in vulnerable people. Mycoplasma was the most abundant of the Mycoplasmatales, and these cause pneumonia in humans.
...So, how does sequencing the bacteriome of wild African buffalo blood help with disease surveillance?
My opinion is that this study is the first step in building a picture of the landscape of potential zoonotic pathogens in a region. What I think is needed now is similar profiling of the microbiome of disease vectors, natural flora, soil and other wildlife in an area/ region to develop complete picture. Eventually, by putting together a complete ‘background’ or‘business as usual’ picture of the microbiome profile of a region. Any deviation from this would quickly flag when there could be an outbreak or emerging pathogen of concern.
It is a big task, of course, and not an easy one…and sounds like something that you may only find in science fiction films/ books at the moment. But it could be possible with the development of new technologies, and collaboration between One Health experts and microbiome researchers.
Follow the Topic
-
BugBitten

A blog for the parasitology and vector biology community.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in