High-quality and robust protein quantification in large clinical/pharmaceutical cohorts with IonStar proteomics investigation
Published in Protocols & Methods

For decades1, quantitative proteomics, i.e. the comparison of the abundances of numerous proteins between biological/clinical groups, has afforded extensive understanding of biological functions, disease mechanisms, and therapeutic effects, and facilitated the discovery/development of new drugs and therapies.
To obtain reliable biological, pharmaceutical, and clinical information by quantitative proteomics, attaining high data quality that meets the following criteria is critical: i) accurate: the method should faithfully reveal the true difference in the abundances of the measured proteins between groups of comparison; ii) reproducible: it should produce the same result in multiple independent analyses, and iii) robust: it should work consistently for all the samples in the cohort.
However, these essential requirements are difficult to meet. For instance, non-specific detection of protein/peptide signals in biological samples represents a common technical challenge, resulting in low accuracy and bad reproducibility in protein quantification, as well as the absence of protein abundance data in some of the samples (i.e. missing values). These issues are much more profound for proteins with relatively low abundances, which are exactly what we are going after, because the low-abundance proteins are most likely those with key biological functions2.
The above problems are manifested further when measuring a large cohort of samples, which are often required for a typical clinical and pharmaceutical investigation. For example, due to the substantial inter-individual variations among patients, a sufficient number of individuals or samples (e.g. >50/group) should be analyzed to gain enough statistical power. Another example is, to evaluate drug efficacy, multiple treatment groups, each with a number of time points after drug dosing, should be investigated, which can easily require the analysis of >100 samples in one experiment.
Compared to the analysis of <20 samples in a typical proteomics quantification experiment, it is far harder to achieve accurate, reproducible quantification of low-abundance proteins across a much larger cohort, particularly, maintaining reasonable data quality from the first to the last sample in the cohort.
The quantitative data with inferior quality not only masks important biological clues but also causes a high false-positive rate in finding the significantly changed proteins between groups, which is critical to discover what a drug or disease does to a biological system. Unfortunately, though the field has been intensively focusing on other metrics such as the number of quantifiable proteins, the lowest amount of samples used, etc., the fundamental importance of high-quality quantification tends to be overlooked3,4.
To address these challenges and achieve accurate, reproducible, and robust proteomic quantification in a large cohort of samples, with years of intensive efforts, we have developed a novel pipeline, named IonStar. The very core feature of this strategy is to quantify proteins by picking up the ultra-high resolution MS1 signals of individual peptides from tissue, cell, or plasma samples, using precisely defined narrow mass windows (Figure 1).
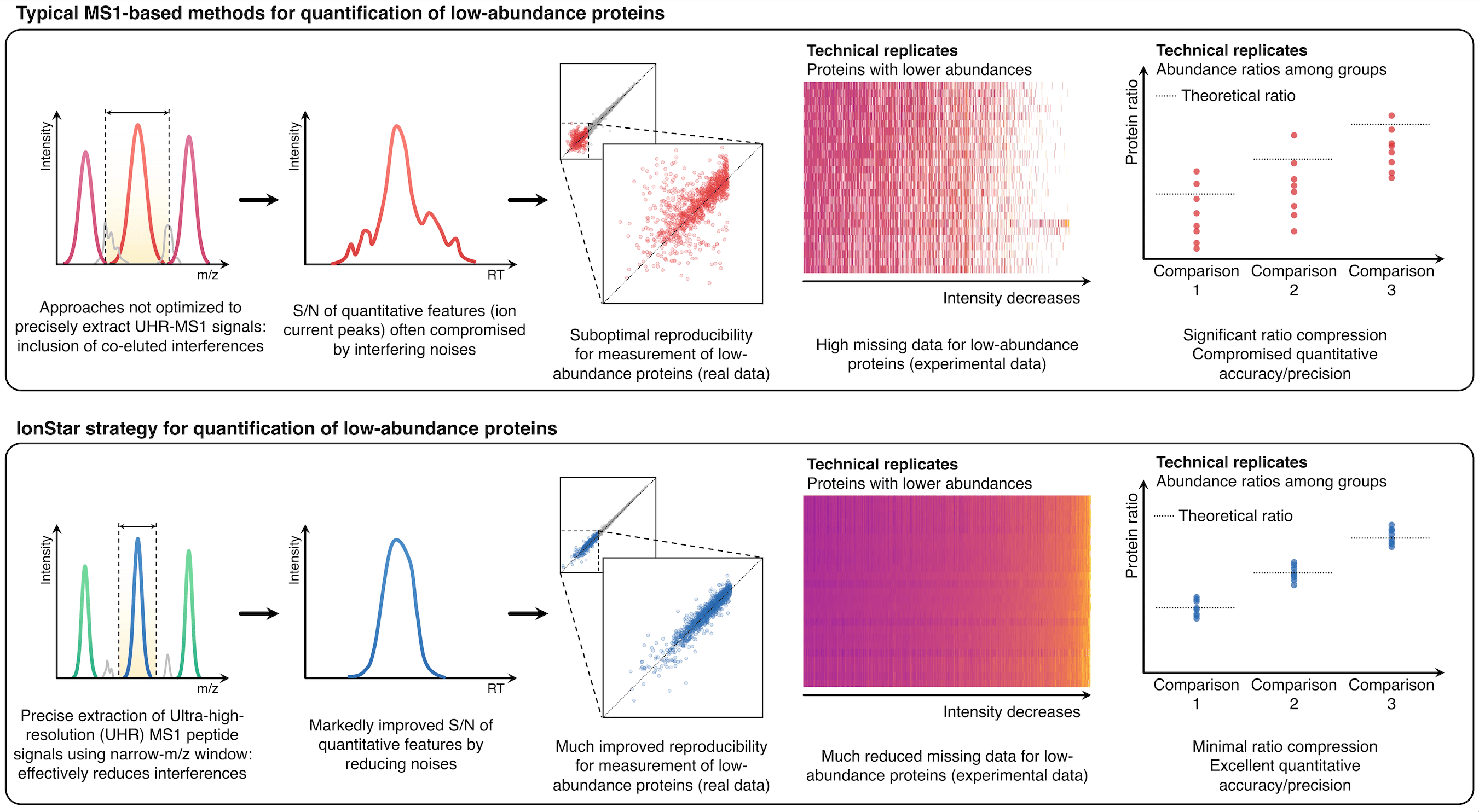
Figure1. IonStar substantially improves the specificity, accuracy, and precision for quantification of low-abundance proteins by taking advantage of the high sensitivity and selectivity attainable by UHR-MS1 (e.g., 120-240k FWHM@m/z=200), which is now widely available on Orbitrap instruments.
This strategy substantially minimizes interfering signals, which is the number one source for low-quality quantitative data, especially for low-abundance proteins, and thus achieves highly selective protein quantification. Additionally, the ultra-high-resolution MS1 signals, when procured properly, are stable and intensive, permitting sensitive, reproducible, and accurate quantification across a large cohort.
Centered on this unique quantification strategy, IonStar has three components that work synergically: i) reproducible and robust sample preparation, ii) extensive, reproducible, and sensitive LC-MS (liquid chromatography-mass spectrometry) analysis, and iii) a data processing pipeline for high-quality quantification of proteins with stringent quality controls (Figure 2). For quantitative proteomics, it is of vital importance to understand that highly robust and reproducible experimental procedures are essential to ensure a top-class quantitative performance, though this aspect has been often overlooked. The optimal sample preparation and LC-MS analysis procedures, as well as the data processing approaches, were developed via years of strenuous efforts on optimization and evaluation (Figure 2). Individually, the sample preparation, LC-MS analysis, and data processing method have all shown superior performance when compared to their “peer” techniques4,5,6,7.
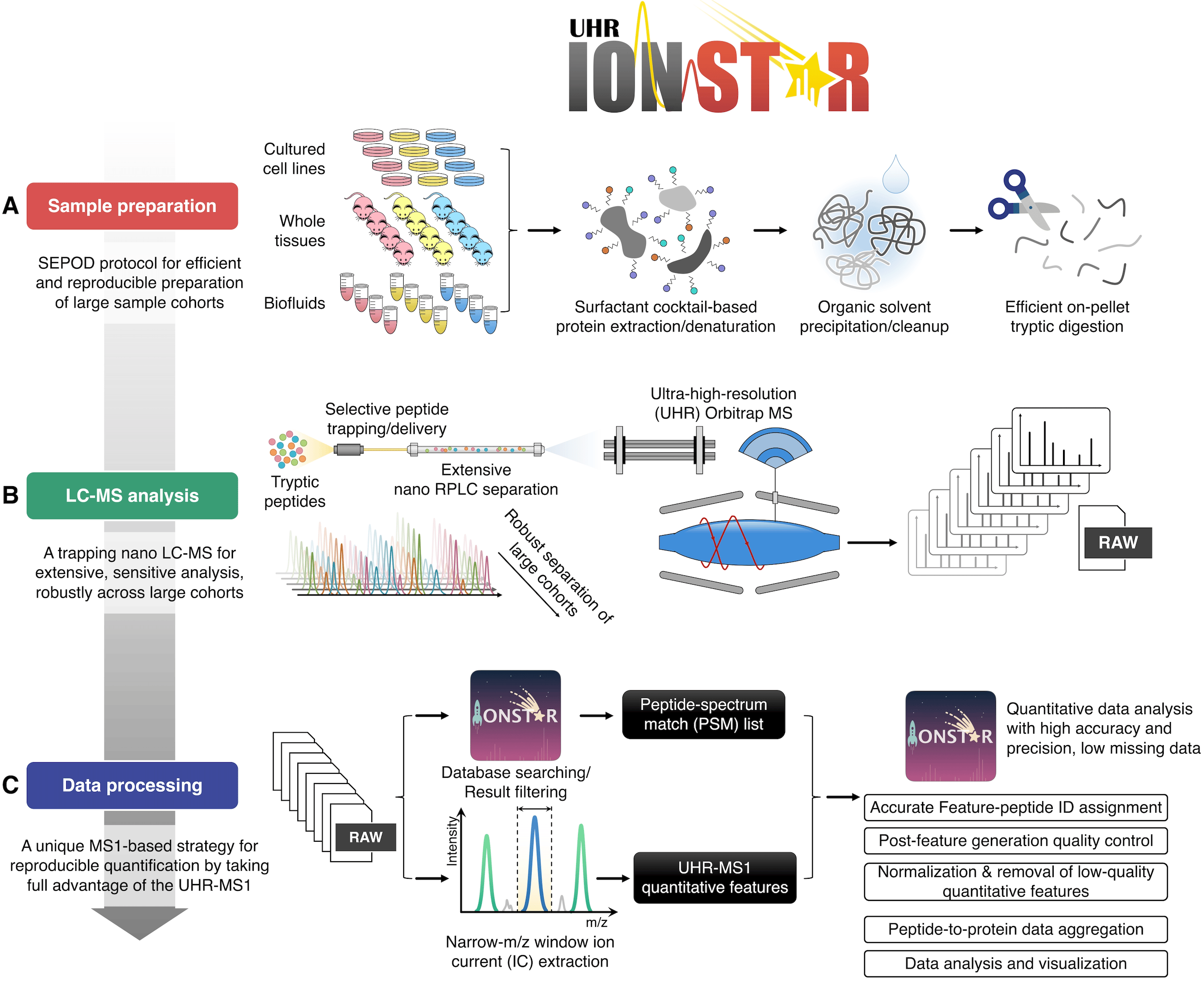
Figure 2. The scheme of the IonStar protocol.
All in all, IonStar is a unique protocol that is designed and optimized for accurate, reproducible, and robust protein quantification in large sample cohorts, and therefore enables high-quality clinical and pharmaceutical investigations. The protocol has been adopted in a large body of publications that lead to insightful findings in clinical and pharmaceutical investigations; it also is well-accepted by many pharmaceutical scientists from the industry. We heartily recommend this protocol to the research community, especially to those who desire high-quality proteomics quantification in clinical and pharmaceutical studies.
References
1 Patterson, S. D. & Aebersold, R. H. Proteomics: the first decade and beyond. Nat Genet 33 Suppl, 311-323 (2003).
2 Gramolini, A., Lau, E. & Liu, P. P. Identifying Low-Abundance Biomarkers. Circulation 134, 286-289 (2016).
3 Wang, X., Shen, S., Rasam, S. S. & Qu, J. MS1 ion current-based quantitative proteomics: A promising solution for reliable analysis of large biological cohorts. Mass Spectrometry Reviews 38, 461-482 (2019).
4 Wang, X. et al. Ultra-High-Resolution IonStar Strategy Enhancing Accuracy and Precision of MS1-Based Proteomics and an Extensive Comparison with State-of-the-Art SWATH-MS in Large-Cohort Quantification. Analytical Chemistry 93, 4884-4893 (2021).
5 Shen, S. et al. Surfactant Cocktail-Aided Extraction/Precipitation/On-Pellet Digestion Strategy Enables Efficient and Reproducible Sample Preparation for Large-Scale Quantitative Proteomics. Anal Chem 90, 10350-10359 (2018).
6 Shen, X. et al. An IonStar Experimental Strategy for MS1 Ion Current-Based Quantification Using Ultrahigh-Field Orbitrap: Reproducible, In-Depth, and Accurate Protein Measurement in Large Cohorts. Journal of Proteome Research 16, 2445-2456 (2017).
7 Shen, X. et al. IonStar enables high-precision, low-missing-data proteomics quantification in large biological cohorts. Proceedings of the National Academy of Sciences 115, E4767-E4776 (2018).
Follow the Topic
-
Nature Protocols

This journal publishes secondary research articles and covers new techniques and technologies, as well as established methods, used in all fields of the biological, chemical and clinical sciences.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in