High-throughput microfluidic device streamlines aging studies in Cryptococcus neoformans
Published in Protocols & Methods
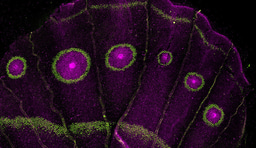
Experimental data with Cryptococcus neoformans infection models indicate that cells of advanced generational age accumulate during chronic infection. More importantly, C. neoformans cells of advanced generational age are significantly more resistant to antifungal medication than their daughter cells. Opposed to Saccharomyces cerevisiae, Cryptococcus neoformans is an encapsulated yeast and a formidable pathogen that kills over 180,000 people worldwide every year. Persistence of disease, despite presumably effective antifungal therapy is not understood. Research on aged yeasts are required to further explore the mechanisms that allow the emergence of antifungal resistance during the process of aging. Until now, aging research with C. neoformans was limited to small numbers because only low-throughput, conventional aging assays were available. In this issue, Orner et al. presents data that suggests these challenges can be overcome by use of a novel microfluidic system coupled to time-lapse microscopy with a chip that is specifically adapted to accommodate aging of C. neoformans.
These advancements with a modified microfluidic platform now permit studies on the marked stochasticity of replicative lifespan in clinical C. neoformans populations. Furthermore, they will also permit the capture of the stochasticity of protein expression in individual cells. Orner et al. presents experiments that study the dependence of generational age and antifungal resistance on a cellular level. Pilot studies with knock out mutants of genes that were found to be significantly upregulated in old cells are performed. These pilot studies have already identified interesting potential drug targets. For example, the knock mutant ∆CNAG_04546, a presumed multi-drug transporter. has a baseline unchanged minimum inhibitory concentration to amphotericin B, a recommended antifungal for this disease. However, once aged on the HYAA chip to 10 generations, this mutant does not acquire the age dependent drug resistance that we see in the wild type strain.
In summary, traditional methods to study aging in C. neoformans were expensive, inefficient, and could not capture variability of phenotypes, which is inherent to pathogen populations exposed to selection pressure in vivo and may drive persistence. Here, we demonstrate the development and use of a High-Throughput Yeast Aging Analysis for Cryptococcus (HYAAC) microfluidic device to improve the study of aging and age-associated genes in C. neoformans. Compared to traditional methods, the HYAAC is superior in its efficiency to isolate, manipulate, and observe old cells for analysis. This novel HYAAC chip will foster research on age-related phenotypes, which may contribute significantly to virulence and cannot be studied in young exponentially grown cell populations.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: Feb 28, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in