Highlights of the BMC Series – July 2025
Published in Healthcare & Nursing, Ecology & Evolution, and Protocols & Methods
BMC Ecology and Evolution - Evolutionary insights into Na+/K+-ATPase-mediated toxin resistance in the Crested Serpent-eagle preying on introduced cane toads in Okinawa, Japan
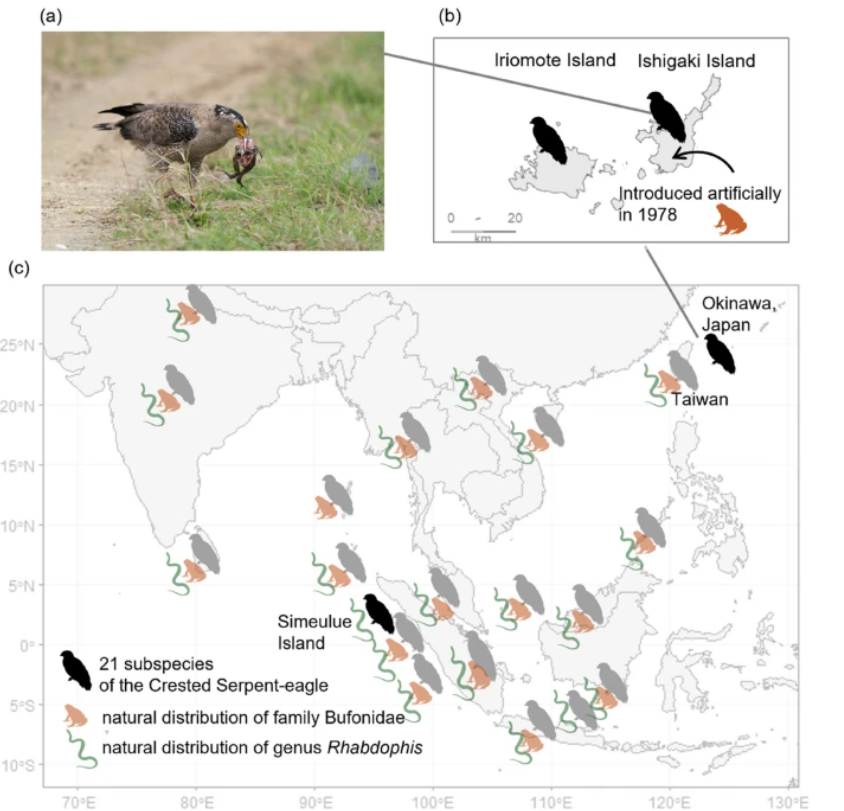
Crested Serpent-eagles on the Japanese islands of Ishigaki and Iriomote prey on cane toads (Rhinella marina), which defend themselves by secreting cardiotonic steroids. However, these eagles appear unaffected by the toads’ toxins, raising questions about whether they have evolved resistance to cardiotonic steroids.
A recent study published in BMC Ecology and Evolution explores, for the first time, the genetic population structure of Crested Serpent-eagles on Ishigaki and Iriomote Islands. The aim was to determine whether the two island populations are genetically distinct and whether they show genetic signatures associated with resistance to cardiotonic steroids. The researchers also investigated the evolutionary history of potential toxin resistance, focusing on the ATP1A1 gene, which encodes a sodium-potassium pump protein—a known target of these toxins. Mutations in this gene have been linked to toxin resistance in other vertebrates.
Whole-genome resequencing revealed that the Ishigaki and Iriomote eagle populations are genetically distinct, and that the Crested Serpent-eagle carries specific substitutions in the ATP1A1 gene which may confer toxin resistance. This potential resistance appears to have been maintained throughout the species’ evolutionary history.
Furthermore, the closely related Black-chested Snake Eagle shares a similar ATP1A1 gene sequence, suggesting that natural selection may have shaped this trait within their lineage.
However, a lack of genetic, molecular, and dietary data on raptors currently limits a full understanding of the evolutionary origins of toxin resistance. The study concludes that further research is needed to clarify the mechanisms underlying toxin resistance in the Crested Serpent-eagle.
BMC Oral Health - Early-life snack and drink consumption patterns among children: findings from a U.S. birth cohort study
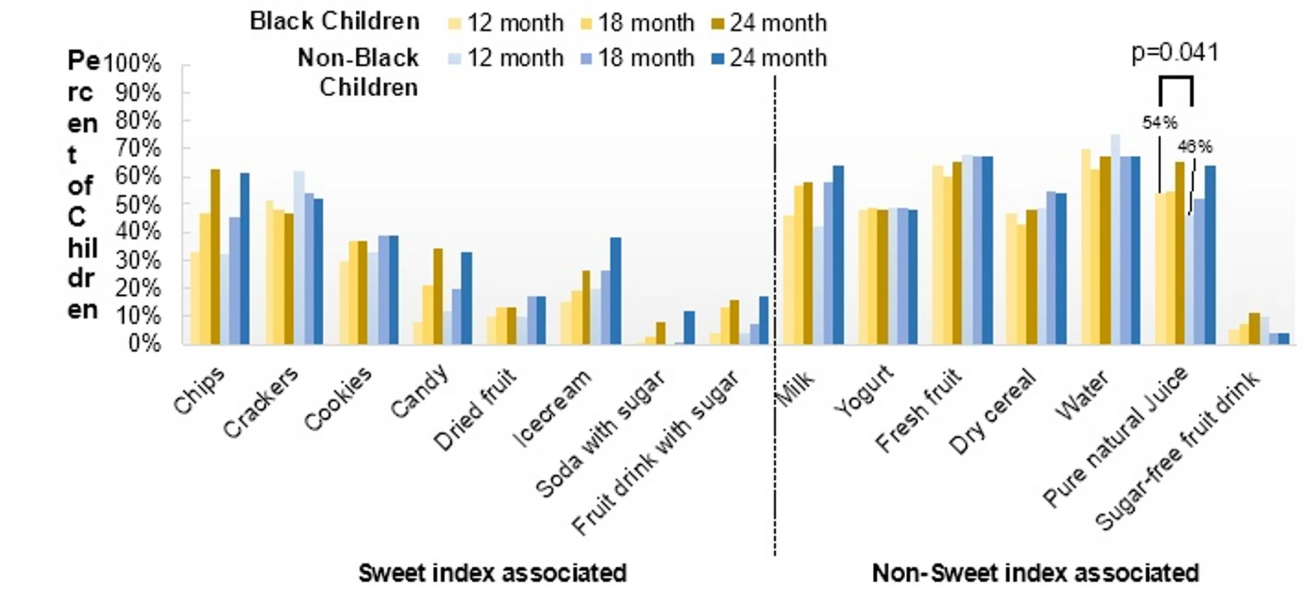
Early childhood diets play a crucial role in long-term oral health, with children from low-income and racially minoritised backgrounds in the U.S. experiencing higher rates of early childhood caries (ECC), often due to frequent consumption of sugary drinks and processed snacks. While it is well established that diet contributes to ECC, key questions remain about how specific snacking and drinking habits influence risk during the earliest years of life.
A recent study published in BMC Oral Health examines dietary patterns during the first two years of life among underserved birth cohorts, with the aim of better understanding these links and informing more targeted, evidence-based strategies to improve oral health equity. To achieve this, researchers analysed a birth cohort of 127 Medicaid-eligible children aged 1 to 2 years, recruited from two university-affiliated clinics in Upstate New York. Parents completed structured dietary questionnaires when their children were 12, 18, and 24 months old, allowing researchers to track the types of snacks and drinks consumed.
Snacks were grouped based on their cavity-causing potential, and Latent Class Analysis (LCA) was used to identify common patterns in snack and drink consumption and to explore how these patterns related to early signs of tooth decay.
The study found that young children from underserved communities exhibit diverse snack and drink consumption behaviours. However, despite identifying distinct dietary patterns, no significant association was observed between these patterns and the onset of ECC—highlighting the multifactorial nature of the condition.
BMC Health Services Research - The meaning of ‘quality’ of homecare for older people: a scoping review
Formal homecare—delivered by paid care workers under regulated systems—plays a vital role in helping older adults live independently in their own homes. These services support daily activities such as washing, cooking, and shopping, and may also extend to helping individuals stay connected with their communities. Unlike unpaid care from family or friends, formal homecare can also indirectly support informal caregivers by easing their burden and helping to sustain crucial care networks.
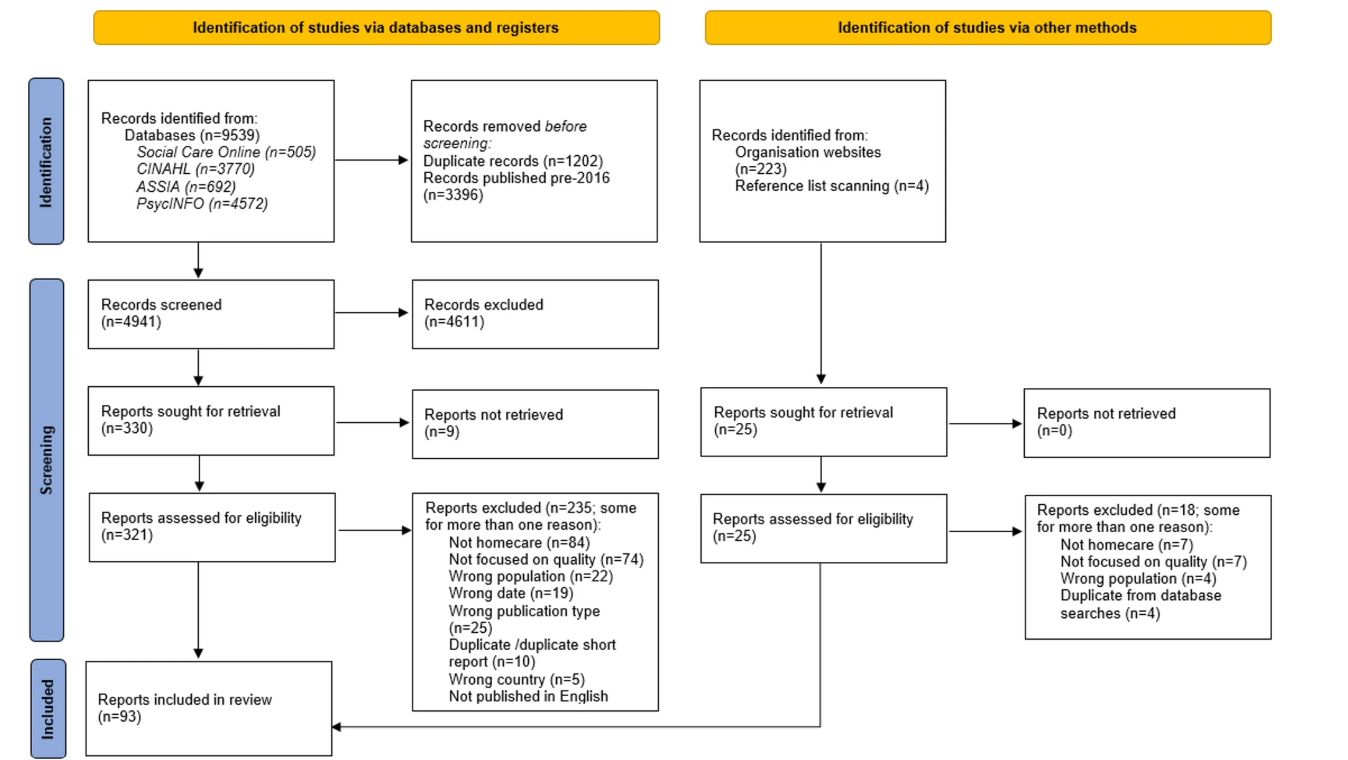
This scoping review, published in BMC Health Services Research, analysed 93 articles from 16 high-income countries to explore how different stakeholders understand the concept of ‘quality’ in homecare for older people. It highlighted both shared and differing perspectives, and identified the qualitative and quantitative measures used to assess quality.
The review found strong agreement across countries and stakeholder groups on the meaning of quality in homecare. Quality is widely seen as a multi-dimensional concept, with particular emphasis on personal relationships and services tailored to individual needs. However, despite recognition of the importance of data in shaping and measuring quality, there is still uncertainty about what types of data should be collected and how best to use them.
To improve care delivery, the authors recommend the development of an internationally agreed, multi-dimensional framework for measuring homecare quality—one that reflects the views of multiple stakeholders while keeping the burden on providers and families manageable.
BMC Methods - Integrated protocol for single-cell RNA sequencing, organoid culture, and RNA-protein spatial analysis in gastroesophageal tissues
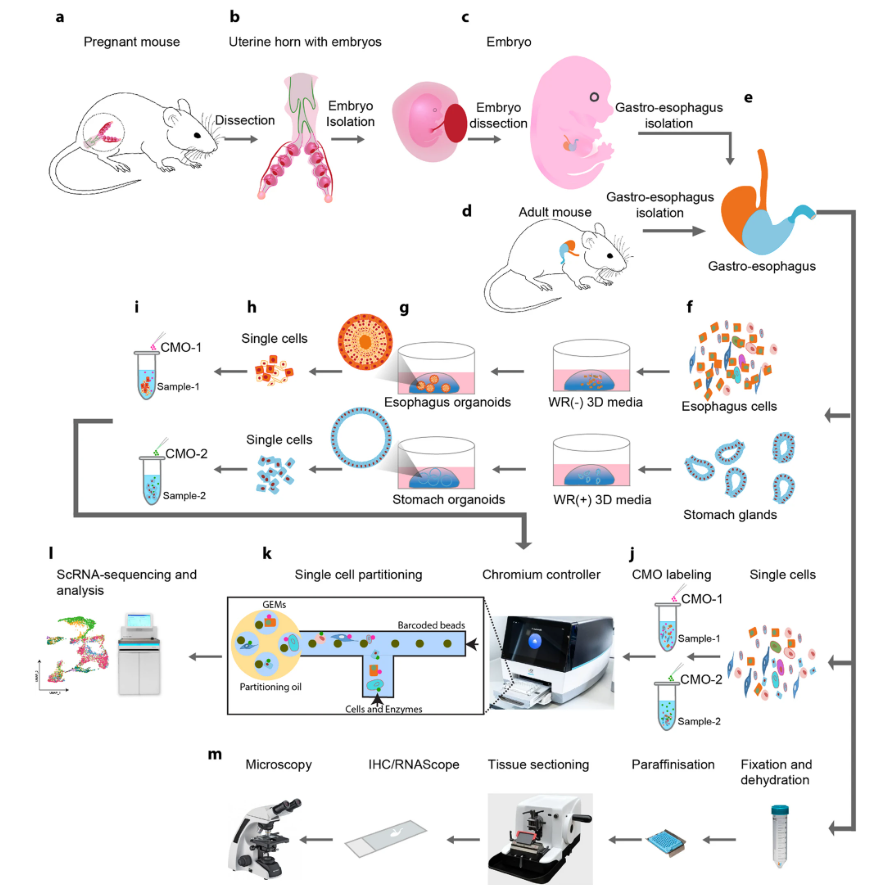
The gastroesophageal tract—which includes the esophagus, gastroesophageal junction (GEJ), and stomach—plays a vital role in digestive health. Disruptions in its cellular and molecular composition can lead to a range of conditions, including gastroesophageal reflux, infection-related inflammation, Barrett’s esophagus, and even cancer. A clear understanding of tissue development and homeostasis is therefore essential to uncover the mechanisms underlying these diseases.
A recent study published in BMC Methods presents a high-quality, integrated protocol that allows researchers to isolate single cells from mouse gastroesophageal tissues—ranging from early embryonic stages to adulthood. This approach enables high-resolution single-cell RNA sequencing (scRNA-seq), offering detailed insight into gene expression and cellular heterogeneity.
In addition to sequencing, the authors describe a method for generating 3D organoids from adult epithelial stem cells. These organoids closely mimic real tissue architecture, making them valuable models for studying development and disease. The study also introduces a spatial analysis workflow that combines single-molecule RNA in situ hybridisation with immunofluorescence, allowing researchers to simultaneously visualise gene activity and protein expression within intact tissue sections.
Together, these tools provide a powerful platform for investigating cellular dynamics, tissue architecture, and disease mechanisms in the gastroesophageal tract.
BMC Artificial Intelligence - Inconsistency detection in cancer data classification using explainable-AI
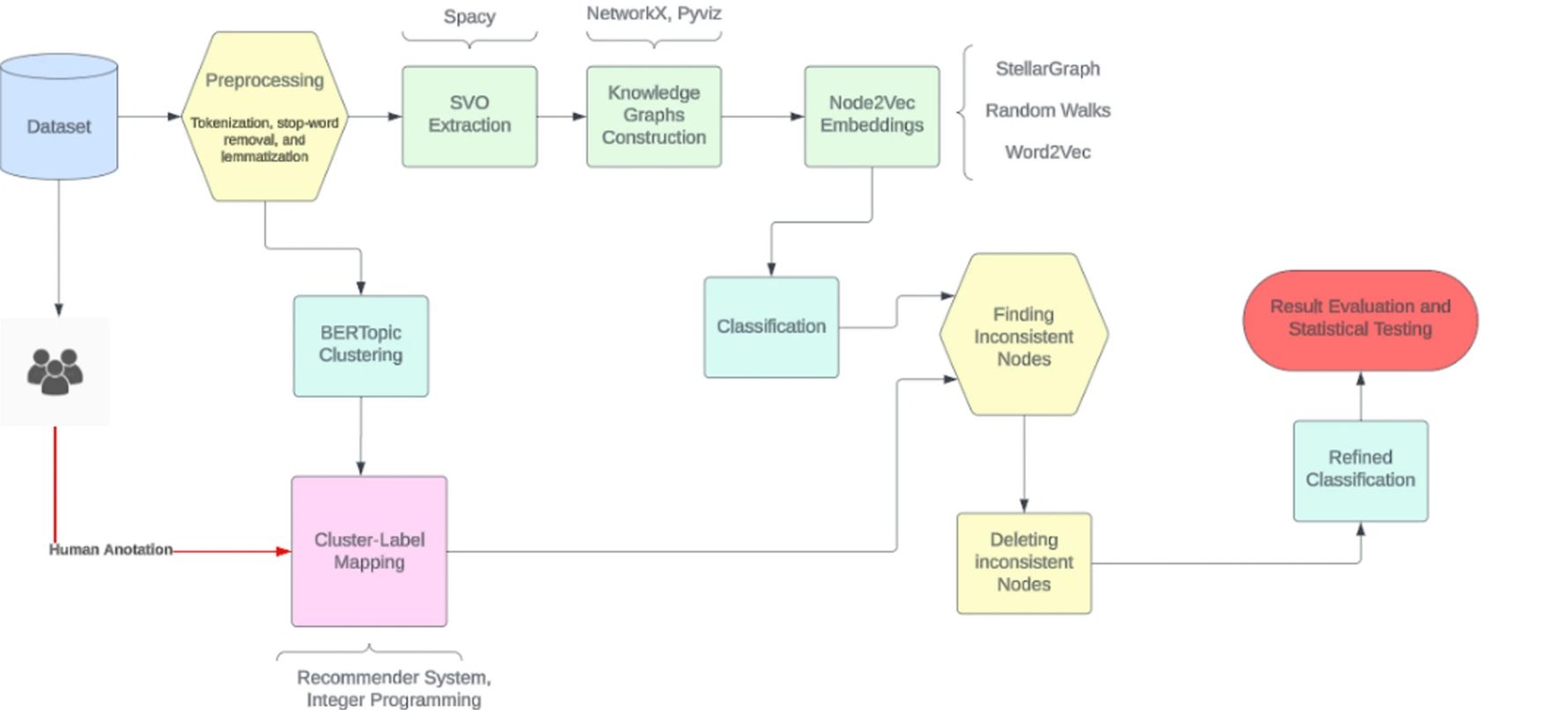
Accurate classification of cancer-related textual data is essential for enabling timely diagnosis and treatment. However, many existing classification methods lack clarity due to data inconsistencies and mislabelling, making it difficult to analyse prediction errors. In addition, manual error analysis is often time-consuming and prone to mistakes.
To address these challenges and systematically identify and analyse errors, a study published in BMC Artificial Intelligence developed and employed explainable AI techniques. The researchers designed a dual-perspective approach that combines two key components: unsupervised semantic clustering and supervised classification.
On one hand, BERTopic (unsupervised clustering) groups texts based on their underlying meaning. On the other, a support vector machine – SVM (supervised classification) classifier uses graph-based embeddings (via Node2Vec) to analyse structural relationships within the data.
The framework also incorporates an Explainable Inconsistency Detection (EID) module, which identifies and eliminates outlier nodes through targeted feature refinement. To enhance the accuracy of label assignments, a collaborative filtering recommender system aligns clusters with the most likely correct labels. Finally, the model’s performance improvements are validated using robust statistical tests.
When tested on cancer datasets, the approach led to notable improvements. Classification accuracy increased from 46% to 91%, and the F1-score rose from 50% to 89%. Importantly, statistical analysis showed that these improvements weren’t due to random data removal but to the targeted identification and exclusion of problematic data points. This work offers valuable insight by combining semantic clustering with structural classification and enhancing explainability through the EID framework.
Follow the Topic
-
BMC Health Services Research

An open access, peer-reviewed journal that considers articles on all aspects of health services research, focusing on digital health, governance, policy, system quality and safety, delivery and access, financing and economics, implementing reform, and the workforce.
-
BMC Oral Health

This is an open access, peer-reviewed journal that considers articles on all aspects of the prevention, diagnosis and management of disorders of the mouth, teeth and gums, as well as related molecular genetics, pathophysiology, and epidemiology.
-
BMC Ecology and Evolution

An open access, peer-reviewed journal interested in all aspects of ecological and evolutionary biology.
-
BMC Methods

An open-access, peer-reviewed journal that focuses on publishing lab protocols and methodology papers in the natural sciences; including biology, chemistry, physics, computational and biomedical sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Bioacoustics and soundscape ecology
BMC Ecology and Evolution welcomes submissions to its new Collection on Bioacoustics and soundscape ecology. By studying how animals use sound and how noise impacts them, you can learn a lot about the well-being of an ecosystem and the animals living there. In support of the United Nations Sustainable Development Goals (SDGs) 13: Climate action, 14: Life below water and 15: Life on land, the Collection will consider research on:
The use of sound for communication
The evolution of acoustic signals
The use of bioacoustics for taxonomy and systematics
The use of sound for biodiversity monitoring
The impacts of noise on animal development, behavior, sound production and reception
The effect of anthropogenic noise on the physiology, behavior and ecology of animals
Innovative technologies and methods to collect and analyze acoustic data to study animals and the health of ecosystems
Reviews and commentary articles are welcome following consultation with the Editor
(Jennifer.harman@springernature.com).
Publishing Model: Open Access
Deadline: Mar 27, 2026
Ecology of soils
BMC Ecology and Evolution invites researchers to submit their work on soil ecosystems and their implications for environmental sustainability. Soils are living, dynamic systems that support a vast array of organisms, from tiny microbes to plants and animals. They play a vital role in keeping our planet healthy by recycling nutrients, storing carbon, and helping regulate water and climate. Understanding how soil life functions and adapts is essential for tackling major environmental challenges like climate change, habitat loss, and soil degradation.
This collection of articles aims to showcase the latest research in soil ecology, emphasizing the interactions among soil organisms, biogeochemical processes, and ecosystem functions across various landscapes. We invite submissions that explore the following topics:
•Microbial and faunal diversity in soils: Patterns, drivers, and functional roles of bacteria, fungi, protists, and invertebrates
•Soil biogeochemistry and ecosystem functioning: Nutrient cycling, carbon sequestration, and soil health in both natural and managed ecosystems
•Soil-plant interactions: Rhizosphere processes, plant-microbe symbioses, and their effects on vegetation dynamics
•Land use and climate change impacts on soil communities: Responses of soil biodiversity and functions to agricultural intensification, urbanization, pollution, and climate shifts
•Soil metagenomics, phylogenetics, and functional ecology: Advances in molecular approaches for studying soil microbial and faunal communities
•Conservation and restoration of soil ecosystems: Strategies for maintaining soil biodiversity and ecosystem services in degraded landscapes
All manuscripts submitted to BMC Ecology and Evolution, including those submitted to collections and special issues, are assessed in line with our editorial policies and the journal’s peer review process. Reviewers and editors are required to declare competing interests and can be excluded from the peer review process if a competing interest exists.
This Collection supports and amplifies research related to SDG 15: Life on Land.
Publishing Model: Open Access
Deadline: Jun 02, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in