HJURP facilitates DNA repair and confers radio-resistance to glioma cells by promoting chromatin reorganization
Published in Cancer, Cell & Molecular Biology, and Genetics & Genomics
Glioblastoma, a highly aggressive form of brain cancer, is notorious for its lethality due to its invasive nature into the surrounding brain tissue and resistance to standard treatments [1]. The resistance of glioblastomas to radiation therapy diminishes treatment efficacy, contributing significantly to the poor prognosis associated with this aggressive brain cancer. Therefore, understanding the mechanisms underpinning glioblastoma’s resistance becomes paramount in the quest for more effective treatment strategies. This challenge motivated our study, aiming to unravel the factors contributing to resistance of glioblastoma cells to radiation therapy and explore potential avenues for overcoming this treatment barrier.
Previous work from our group identified significantly elevated levels of the Holliday Junction Recognition Protein (HJURP) in glioblastoma compared to low-grade gliomas or normal tissues [2-3]. HJURP has emerged as a key player in several cancer types, demonstrating a strong correlation with patient survival [2-11]. Despite this association, the mechanistic bases of HJURP involvement in cancer aggressiveness have remained elusive. Our study uncovers the intricate roles of HJURP, shedding light on its impact on chromatin structure, DNA repair, and potential as a therapeutic target for glioblastoma.
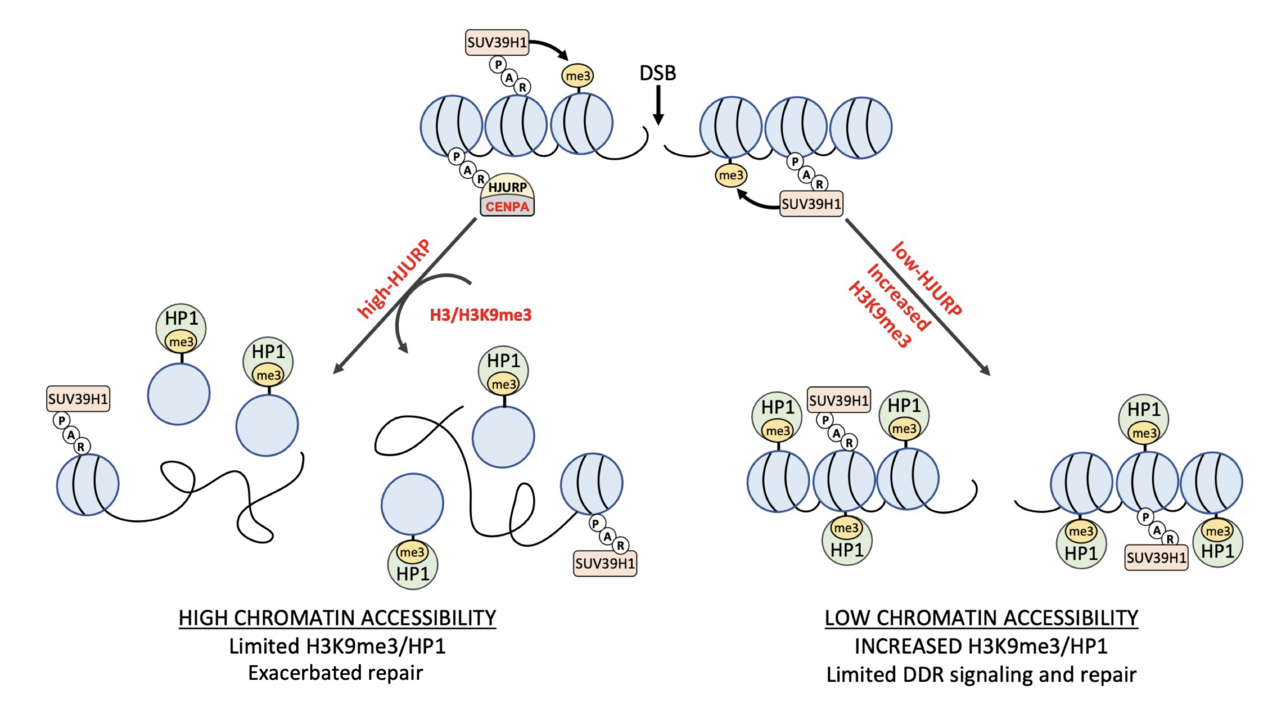
At the molecular level, HJURP orchestrates the loading of the histone H3 variant, CENP-A, at the centromeric chromatin [12-14]. This action epigenetically defines centromeres, which are crucial for proper chromosome segregation. Additionally, HJURP exhibits associations with DNA repair, a facet of its function that has been minimally explored until now. Our research demonstrated that HJURP is recruited to DNA double-strand breaks (DSBs) through a mechanism reliant on chromatin PARylation. Once at the DSB sites, HJURP instigates epigenetic alterations to drive efficient DNA repair. Its presence promotes the turnover of the repressive chromatin components, H3K9me3 and HP1, facilitating DNA damage signaling and aiding in DSB repair especially through the homologous recombination pathway.
Furthermore, HJURP's influence extends beyond DNA damage response, as its overexpression in glioma cell lines induces a global restructuring of heterochromatin, even in the absence of DNA damage. This not only alters DNA damage signaling but also enhances the radio-resistance of glioma cells. Crucially, our findings indicate that the levels of HJURP expression in tumors correlate with the poor response of patients to radiation therapy. These data position HJURP as a promising target for the development of adjuvant therapies, offering the potential to sensitize tumor cells to irradiation.
In summary, our study expands the understanding of HJURP's multifaceted roles in DNA repair and chromatin dynamics, presenting it as a compelling candidate for future therapeutic interventions aimed at enhancing the efficacy of cancer treatments. Unraveling the intricacies of glioblastoma resistance is not just a scientific pursuit, it is also a crucial step toward advancing in treatment modalities and offering better therapeutic options to those confronting this disease.
References
1. Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60(3):166-93.
2. Valente V, Teixeira SA, Neder L, Okamoto OK, Oba-Shinjo SM, Marie SK, et al. Selection of suitable housekeeping genes for expression analysis in glioblastoma using quantitative RT-PCR. BMC Mol Biol. 2009;10:17.
3. Valente V, Serafim RB, de Oliveira LC, Adorni FS, Torrieri R, Tirapelli DP, et al. Modulation of HJURP (Holliday Junction-Recognizing Protein) levels is correlated with glioblastoma cells survival. PLoS One. 2013;8(4):e62200.
4. Kato T, Sato N, Hayama S, Yamabuki T, Ito T, Miyamoto M, et al. Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 2007;67(18):8544-53.
5. Hu Z, Huang G, Sadanandam A, Gu S, Lenburg ME, Pai M, et al. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res. 2010;12(2):R18.
6. Li L, Li X, Meng Q, Khan AQ, Chen X. Increased Expression of Holliday Junction Recognizing Protein (HJURP) as an Independent Prognostic Biomarker in Advanced-Stage Serous Ovarian Carcinoma. Med Sci Monit. 2018;24:3050-5.
7. Wang CJ, Li X, Shi P, Ding HY, Liu YP, Li T, et al. Holliday junction recognition protein promotes pancreatic cancer growth and metastasis via modulation of the MDM2/p53 signaling. Cell Death Dis. 2020;11(5):386.
8. Chen T, Zhou L, Zhou Y, Zhou W, Huang H, Yin S, et al. HJURP Promotes Epithelial-to- Mesenchymal Transition via Upregulating SPHK1 in Hepatocellular Carcinoma. Int J Biol Sci. 2019;15(6):1139-47.
9. Wei Y, Ouyang GL, Yao WX, Zhu YJ, Li X, Huang LX, et al. Knockdown of HJURP inhibits non-small cell lung cancer cell proliferation, migration, and invasion by repressing Wnt/beta-catenin signaling. Eur Rev Med Pharmacol Sci. 2019;23(9):3847-56.
10. Chen T, Huang H, Zhou Y, Geng L, Shen T, Yin S, et al. HJURP promotes hepatocellular carcinoma proliferation by destabilizing p21 via the MAPK/ERK1/2 and AKT/GSK3beta signaling pathways. J Exp Clin Cancer Res. 2018;37(1):193.
11. Kang DH, Woo J, Kim H, Kim SY, Ji S, Jaygal G, et al. Prognostic Relevance of HJURP Expression in Patients with Surgically Resected Colorectal Cancer. Int J Mol Sci. 2020;21(21).
12. Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137(3):485-97.
13. Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137(3):472-84.
14. Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, et al. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194(2):229-43.
Follow the Topic
-
Oncogene

This journal aims to make substantial advances in our knowledge of processes that contribute to cancer by publishing outstanding research.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in