How Cells Use DNA Methylation to Know About Their Neighbors: Our Story of a Scientific Discovery on a Monday Morning
Published in Bioengineering & Biotechnology, Protocols & Methods, and Cell & Molecular Biology

Picture your body as a blueprint-driven smart home with an advanced lighting system. This system can finetune the brightness (gene expression) in every room (cell) of the house. DNA methylation plays a pivotal role in this infrastructure by acting as a toggle switch. Responsible for shaping gene expression programs and genome architecture, DNA methylation is integral to numerous biological processes, spanning from embryonic development to disease pathogenesis1, 2. Yet, what precisely regulates this molecular switch? Our research, published in Communications Biology3, unveils a previously overlooked aspect: the involvement of cell-cell junctions in this intricate process.
Our journey began with an unexpected observation. One mundane Monday, during a routine checkup in our laboratory, we observed something odd - our reporter cells, engineered to emit fluorescence as an indicator of DNA methylation - were inexplicably dim! Drawing from literature insights, we were actively using this reporter in conjunction with perturbations of candidate cellular signaling pathways to identify novel regulators of DNA methylation. The idea that a single dish of cells, neglected over a long weekend, could yield anything of value seemed far-fetched. However, even if this observation was unexpected and did not align with our initial experimental path, we chose to explore it further. Little did we realize, that this decision would mark the first step of a journey that would ultimately result in the discovery of a new pathway at the intersection of epigenetics and cell signaling (Figure 1).
A biosensor of DNA methylation helps uncover a new cell signaling pathway
DNA methylation is non-randomly distributed throughout our genome and is heavily concentrated in repetitive elements. These regions need to stay inactive to avoid genomic instability4. Yet, there are times, such as in the preparation of cell division, when controlled activation is necessary. Despite the fact that repetitive DNA comprises around one half of our genome, the cellular signals that are in place to control the DNA methylation landscape at these sites are not clear. This is in part due to methodological challenges associated with the repetitive nature of this DNA, making these regions notoriously difficult to study.
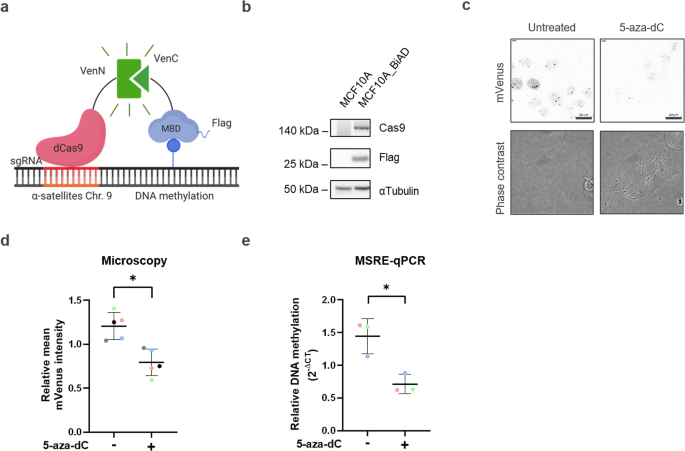
Our breakthrough leveraged a CRISPR/dCas9 fluorescence biosensor we previously developed5 and which enabled us to directly visualize the methylation of repetitive elements within living cells. Systematic experiments with epithelial cell cultures at varying densities and the use of various readouts, confirmed our initial findings: the culture density did influence the DNA methylation levels at α-satellite repeats, the sequences monitored by our sensor. Specifically, in densely populated cultures, like the ones we observed on that Monday morning, the DNA methylation levels of these repeats were lower than under sparse cultivation conditions (Figure 1).
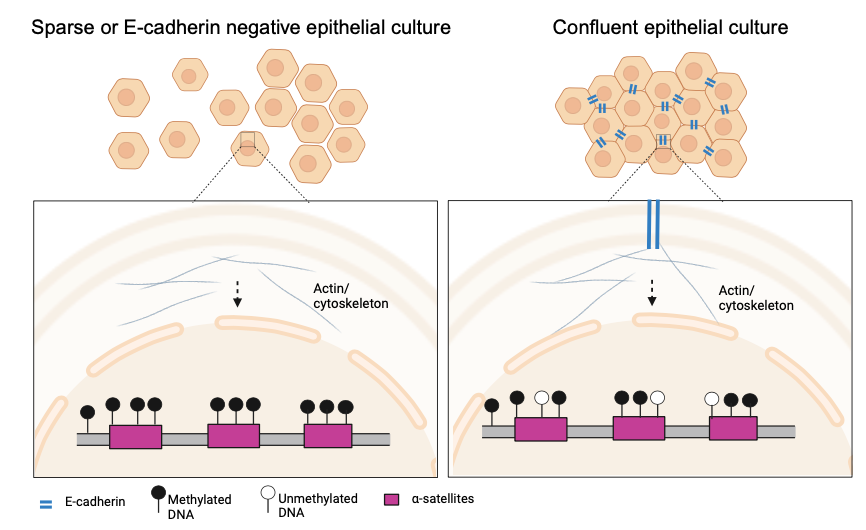
Repeat DNA methylation is modulated by adherens junction signaling
Delving deeper, we turned our attention to the role of cell-cell interactions in this process by focusing on the transmembrane protein E-cadherin. E-cadherin helps form adherens junctions, enabling epithelial cells to stick together. Our experiments showed that disrupting the function of E-cadherin, either through genetic knockdown or by blocking its function, resulted in a marked increase in DNA methylation at the α-satellite repeats. This required an intact actin cytoskeleton. Notably, this mechanism was absent in cancer cells lacking E-cadherin expression and the proliferation of which is not inhibited by cell-cell contacts.
The ramifications of our findings are profound, suggesting that the DNA methylation status of repeats carries information regarding the proximity of neighboring epithelial cells. In densely packed cellular environments, like in a mature epithelial layer, subtle changes in the DNA methylation levels of repetitive elements could thereby be a form of genetic response to cellular crowding. This could play a role in maintaining tissue homeostasis, potentially by coordinating cellular proliferation.
Conclusion: embracing the unexpected to chart new ground
Our journey from a serendipitous observation to uncovering a novel biological insight captures the essence and unpredictability of scientific exploration. It serves as a reminder of the importance of a watchful eye, an open mindset and the value in pursuing unexpected findings, even if these lie outside the intended path. This discovery was achieved through the collaborative efforts of three research groups across two continents, employing a multidisciplinary approach that spans biochemistry, synthetic biology, cell biology, and cell mechanics. Our findings offer a novel perspective on how epithelial cells communicate with one another and shed new light on the intricate relationship between cell signaling and epigenetic pathways. This has significant implications for our understanding of cancer biology and beyond.
For the future, it will be interesting to explore how the DNA methylation machinery is specifically regulated by this newly identified signaling pathway. Additionally, understanding the evolutionary implications of repeat DNA methylation as a mechanism for epithelial cells to sense their neighbors, could reveal how cellular communities have evolved means to maintain tissue integrity and adapt to environmental cues.
References:
- Janssen, S. M., & Lorincz, M. C. (2022). Interplay between chromatin marks in development and disease. Nature reviews. Genetics, 23(3), 137–153. https://doi.org/10.1038/s41576-021-00416-x
- Parry, A., Rulands, S., & Reik, W. (2021). Active turnover of DNA methylation during cell fate decisions. Nature reviews. Genetics, 22(1), 59–66. https://doi.org/10.1038/s41576-020-00287-8
- Brenner, L. M., Meyer, F., Yang, H., Köhler, A. R., Bashtrykov, P., Guo, M., Jeltsch, A., Lungu, C., & Olayioye, M. A. (2024). Repeat DNA methylation is modulated by adherens junction signaling. Communications biology, 7(1), 286. https://doi.org/10.1038/s42003-024-05990-4
- Pappalardo, X. G., & Barra, V. (2021). Losing DNA methylation at repetitive elements and breaking bad. Epigenetics & chromatin, 14(1), 25. https://doi.org/10.1186/s13072-021-00400-z
- Lungu, C., Pinter, S., Broche, J., Rathert, P., & Jeltsch, A. (2017). Modular fluorescence complementation sensors for live cell detection of epigenetic signals at endogenous genomic sites. Nature communications, 8(1), 649. https://doi.org/10.1038/s41467-017-00457-z
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
From RNA Detection to Molecular Mechanisms
Publishing Model: Open Access
Deadline: May 05, 2026
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: May 31, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in