How did Neanderthals regulate their genes?
Published in Ecology & Evolution
Original article here
By Laura Colbran and Tony Capra
“Oh yes, the past can hurt, but the way I see it you can either run from it or learn from it.” – Rafiki, The Lion King
Diverse
evolutionary forces acted on our ancestors, and those past pressures
shaped what humans look like now, including our risk for various
diseases. Our lab studies these evolutionary phenomena from many
different perspectives, ranging from analyzing the evolution of 3D
structures of proteins, to tracing the mechanics of gene regulation
across species, to combining ancient DNA and hospital records to
identify traits affected by genetic variants with interesting
histories.
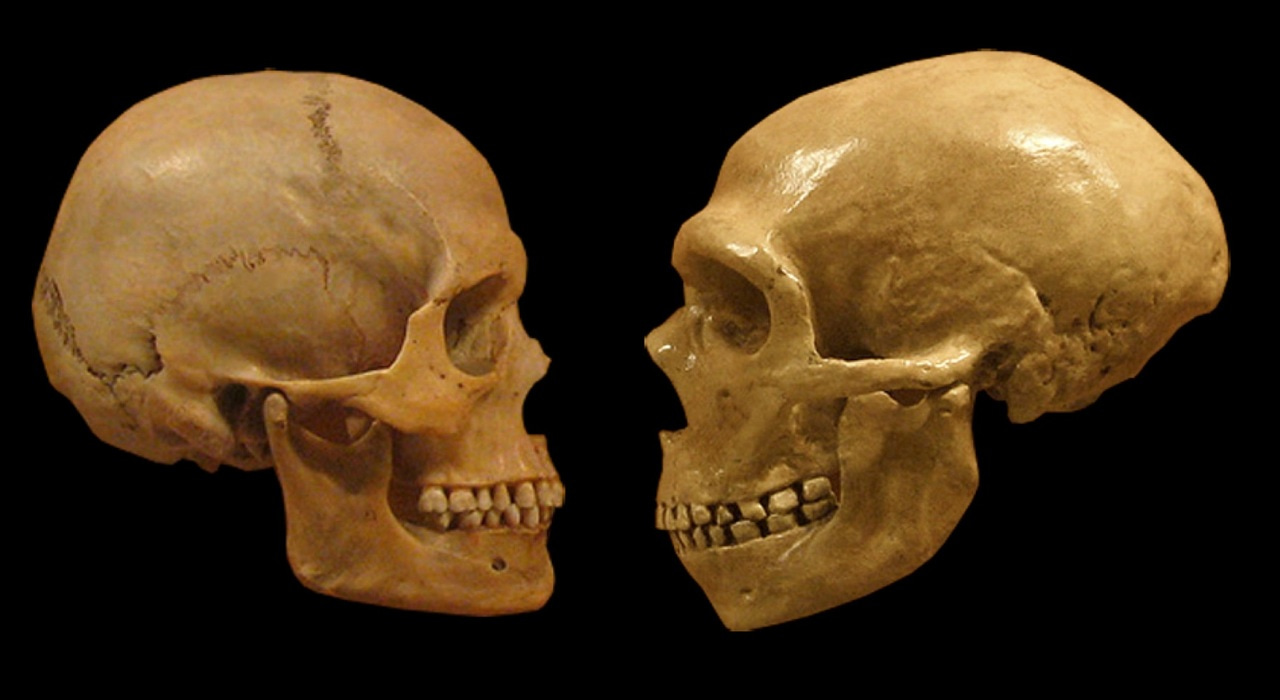
In this study, we were focused on discovering differences between modern humans and our close evolutionary relatives, the Neanderthals and Denisovans (aka “archaic hominins”). Most of what we know about
how we are different comes from studying their bones, but there’s much that these can’t tell us about attributes of whole living organisms. That changed about 10 years ago with the determination of the genome sequence of a Neanderthal who lived tens of thousands of years ago. We now have genomic data from several additional Neanderthals and Denisovans. These new data enable an entirely new kind of comparison between Neanderthals and modern humans—one at the level of individual DNA base pairs. These comparisons revealed
many surprising findings, including that around 2% of the genomes of modern Eurasians comes from Neanderthals due to interbreeding about 40,000 years ago.
From
our work and those of many other labs, we know that most genetic
variants important to recent human evolution and modern disease risk
don’t change genes themselves, but rather influence gene
regulation—when, where, and how much genes are turned on. However,
working out how a genetic variant affects gene regulation and what
repercussions that might have for the organism is difficult, even
when we are studying living humans and can measure gene expression
directly. The problem is even more difficult when studying archaic
hominins who have been dead for tens of thousands of years because
the RNA needed to measure gene expression and the softer tissues to
needed to measure other traits is not preserved with the bones.
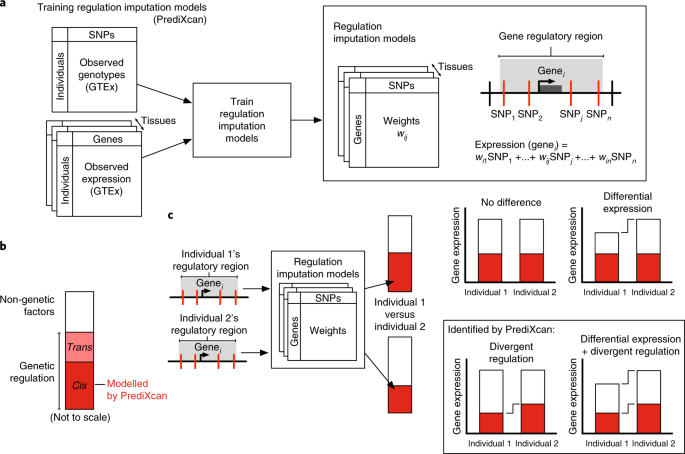
Luckily for us, we hang out with a brilliant group of geneticists who recently developed a computational technique that we thought might be able to help. The technique, called PrediXcan, builds a statistical model that predicts the patterns of gene regulation in a person based only on their combinations of genetic variants.
We
were interested in understanding how gene regulation differed between
archaic hominins and modern humans, and what these differences could
tell us about archaic hominins themselves. Thus, we wondered if we
could apply the PrediXcan approach to archaic genomes.
This was a very exciting possibility, but there was a potential problem. PrediXcan models are trained using genetic information from modern human populations, and while archaic hominins are closely related to modern humans, they evolved separately from us for hundreds of thousands of years. Thus, we weren’t sure that these models would be able to tell us anything useful. But how could we test them? We don’t know the right “answer” for any of the archaic genomes. As a start, we applied the models to modern populations that weren’t included in the training population and showed that they were still useful there. This was promising, but it wasn’t quite satisfying, since archaic hominins are more diverged from modern humans than modern populations are from each other.
Then, we realized that we could measure gene regulatory effects of archaic hominin sequences in some situations. Since Neanderthals interbred with the ancestors of modern Eurasians, Neanderthal variants are present around many genes in living people, and we can measure how they influence gene regulation, albeit in a human context. We took advantage of this to evaluate the accuracy of our approach by retraining prediction models for each gene using only people without Neanderthal ancestry nearby the gene. We then applied these retrained “human-only” models to people who had Neanderthal ancestry near those genes, which was a much closer approximation to applying them to an actual Neanderthal. This experiment worked better than we expected and showed that we could trust many PrediXcan models for our study.
Applying the models to Neanderthal and Denisovan genomes revealed many differences in how genes were regulated between archaic hominins and modern humans. The genes affected were involved in many immune, skeletal, reproductive, and cardiovascular traits. Some of these effects might have been harmful, while others might have been beneficial. To explore these potential effects, we applied our method to more than 20,000 people with DNA information linked to electronic health records. We found that having Neanderthal-like gene regulation sometimes increased risk for disease and sometimes decreased it.
Our work provides a starting point for understanding what specific genes and traits were regulated differently among archaic hominins and thus may be important for being human, as opposed to Neanderthal. As a next step, we are applying our approach to DNA from ancient humans from different times and locations to trace more recent changes in how our genes were regulated. This will further reveal how evolutionary events in our past helped shape modern populations.
Follow the Topic
-
Nature Ecology & Evolution

This journal is interested in the full spectrum of ecological and evolutionary biology, encompassing approaches at the molecular, organismal, population, community and ecosystem levels, as well as relevant parts of the social sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Biodiversity and ecosystem functioning of global peatlands
Publishing Model: Hybrid
Deadline: Jul 27, 2026
Understanding species redistributions under global climate change
Publishing Model: Hybrid
Deadline: Jun 30, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
Terrific work!
Great work indeed! Also a great educative example how maths are strongly related with biology research nowadays...