How the Serval gets its spots
Published in Ecology & Evolution

Discussion of leopard spots and zebra stripes is rarely complete without references to absurd whimsy (Rudyard Kipling) and innovative theory (Alan Turing). While the two authors could not be farther apart in their approach, together they represent the challenge of how one investigates a natural phenomenon in the absence of a suitable experimental system. From appendages to the brain to the skeleton, most aspects of mammalian development can be and have been profitably investigated in laboratory mice, coupling the awesome power of forward genetics with the sophistication of gene-editing. But that's not the case for periodic color patterns—the regularity and complex arrangement of alternating patches of light and dark hair in a cheetah, jaguar, or ocelot have no real counterpart in model organisms.
My research group spent the better part of two decades studying mouse coat color genetics as a model system to understand gene action and interaction. For much of the twentieth century, mouse coat color was to a mammalian geneticist what peas were to Mendel, helping to uncover fundamental paracrine signaling systems that affect body weight, blood cell development, and intestinal ganglia in addition to hair color (1, 2). The advent of next gen sequencing spurred us to think more creatively about color variation in non-model organisms, asking if certain aspects of that variation might inform our understanding of mammalian development and physiology more broadly. For the most part, the genes that control simple color variation like albinism, melanism, and piebaldism are the same in all mammals—studying black leopards or jaguars is just like studying black mice or rats. The biology underlying mammalian color pattern, however, has been an unsolved mystery, one in which we have now gained new insight using domestic cats.

Figure 1. Conceptual framework for mammalian color pattern. Tabby stripes are evident in a fetal cat (a) as soon as pigment is produced, indicating that pattern establishment occurs early in fetal development. After birth, a newborn cheetah (b) and other felids have the same number of dark spots spots as adults; throughout postnatal animal growth and hair recycling, the dark spots increase in size but do not change in number or position; thus, pattern element identity is maintained by permanent cells within the hair follicle. Different types of pigment in hairs of a cheetah (c) and other felids are synthesized by intrafollicular melanocytes in response to paracrine signals from the dermal papilla. tion
We knew from prior work that the spatial information underlying mammalian color pattern is present early in development, long before pigmentation becomes apparent (3). Acquisition of this spatial information, or pattern establishment, is likely to involve alternating groups of embryonic cells that express different groups of genes, a so-called self-regulating "pre-pattern" that develops via a reaction-diffusion mechanism. This idea was conceived of by Alan Turing in 1952, and explored extensively via mathematical modeling and simulation as a theoretical explanation for animal color patterns (4–6). A common feature of reaction-diffusion models is local enhancement and long-range inhibition by a diffusible activator and inhibitor, respectively (7), but the identity and action of the molecules underlying and participating in a putative molecular pre-pattern for cheetahs, jaguars, or other mammals was unknown. Chris Kaelin and Kelly McGowan, who led and carried out all the work in the paper, hypothesized that should such a pre-pattern exist, it might be revealed by single cell RNA-sequencing—clusters of gene expression differences associated with spatially distinct groups of fetal skin cells, but there were two important hurdles: we didn't know the precise time during fetal development when such a pre-pattern might exist, and we didn't have any fetal cat skin!
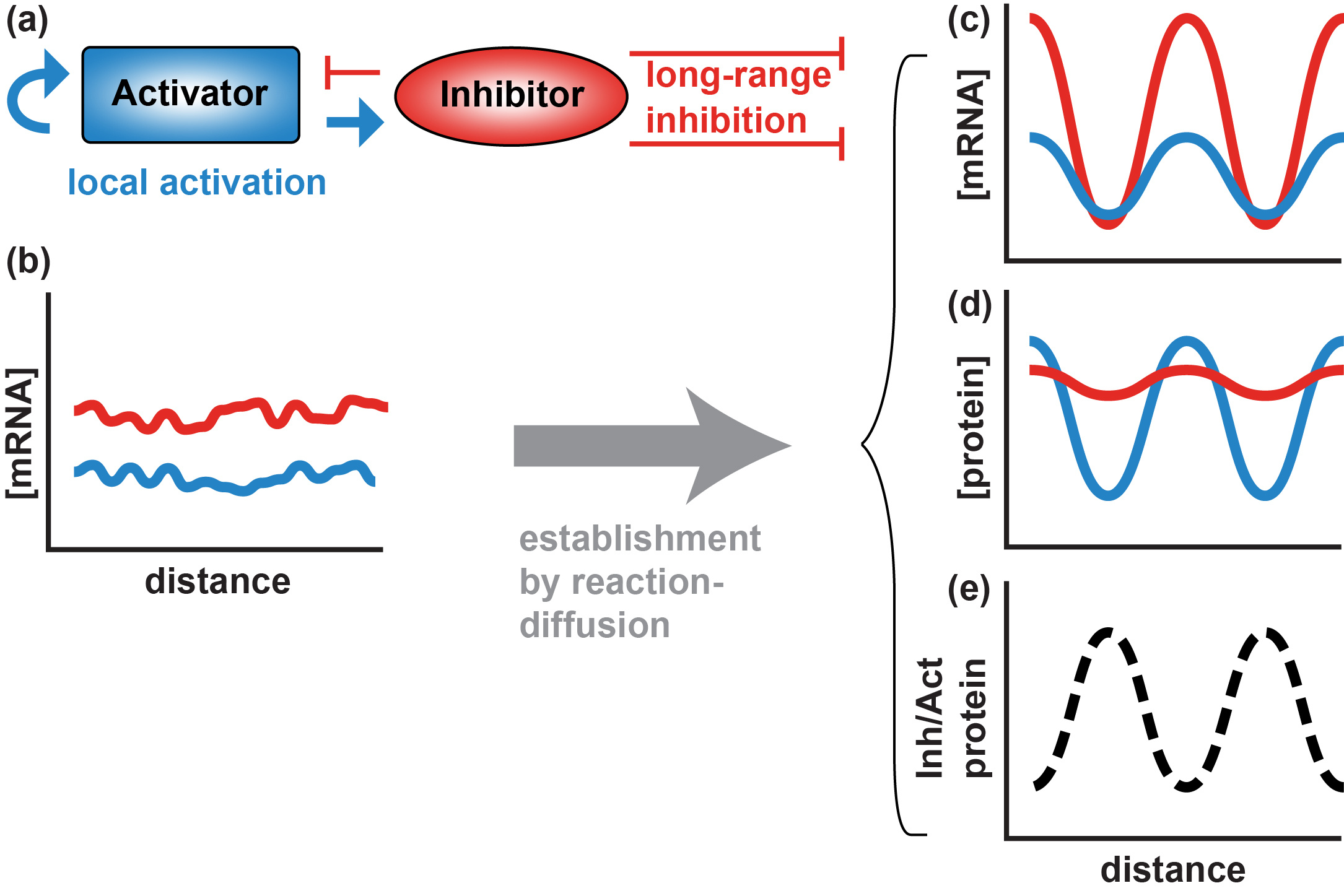
Figure 2. Essential components of a Turing reaction-diffusion model. (a) Hypothetical signals from a diffusible activator with a short radius of action and a diffusible inhibitor with a longer radius of action. Within a field of developmentally identical cells, differences in diffusion coefficients and/or extracellular stability of activator and inhibitor creates conditions in which stochastic variation in expression (b) develops into a self-organizing periodic pattern of mRNA (c) and protein (d), and the ratio of inhibitor to activator protein is high in cells where mRNA expression is low (e). |
 |
The latter hurdle was overcome when Chris and Kelly developed partnerships with several feral cat trap-neuter-release programs, collecting fetal tissue that would otherwise be discarded during spay procedures. Kelly, who believes in the awesome power of careful phenotyping, discovered that during the earliest aspects of skin development before hair follicles are formed, the epidermal layer temporarily exhibits alternating patches of cell columns, which she named "thick" and "thin". Belying the prosaic name, this aspect of skin development had never been described in any mammal. More importantly, the pattern of thick and thin patches in fetal skin predicts the arrangement of colored hairs that would later develop after birth, revealing the developmental window and cell type in which color pattern establishment occurs.
That set the stage for Chris, who believes in the awesome power of genomics, to apply single cell RNA-seq to fetal epidermis just prior to the time at which the thick and thin patches became apparent. The data and analyses of those efforts led to the discovery of a molecular pre-pattern in which a key paracrine signaling molecule, Dickkopf 4 (Dkk4), is produced by cells that develop into thick patches. Fetal expression of Dkk4 marks groups of skin cells that will later harbor patches of dark hair. The family of Dickkopf proteins are inhibitors of Wnt signaling (8, 9), leading Chris and Kelly to propose that Dkk4 serves as a "long-range inhibitor" in a Turing reaction-diffusion model. Our analysis of differentially expressed genes in the thick and thin patches is consistent with this proposal and leads us to predict that genetic and evolutionary alterations that contract or extend the radius of Dkk4 action underlie the different sizes and shapes of color pattern elements in other felids, from cheetahs to servals.
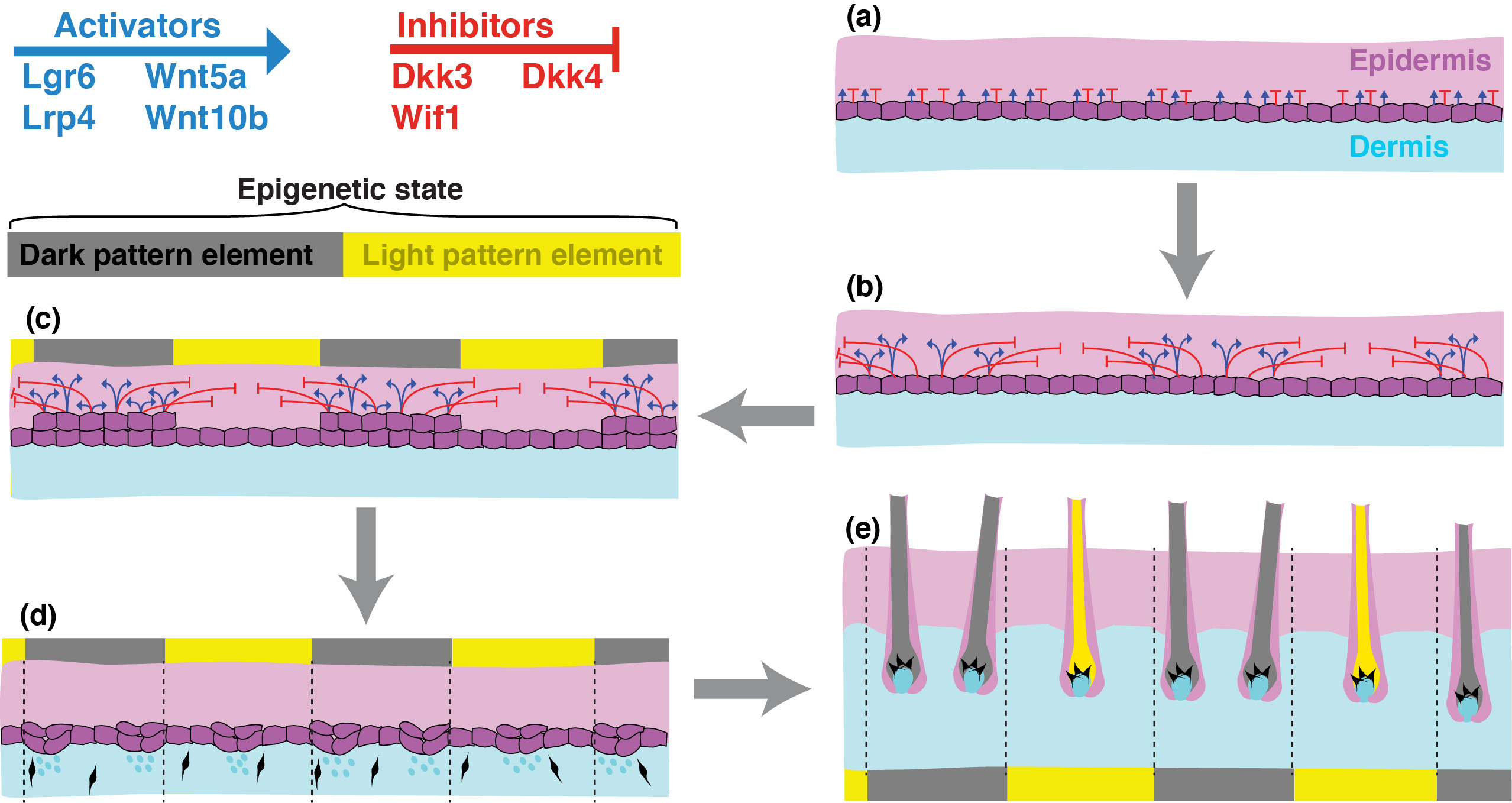
Figure 3. A Turing mechanism for felid color pattern. Single cell RNA-seq analysis of fetal cat skin nominates several candidate molecules as diffusible activators or inhibitors. Stochastic variation of their mRNA levels in basal epidermal cells (a) leads to a self-organizing molecular prepattern during pattern establishment (b), in which the groups of cells that express higher levels of activator and inhibitor mRNA exhibit transient thickening due to local proliferation (c). The molecular prepattern recedes by the time hair follicle formation commences (d), but the sam groups of cells will eventually harbor dark hair, epigenetically maintaining their pattern element identity throughout subsequent growth, development, and hair cycling. Visible evidence of pattern element identity (e) becomes apparent when hair follicle melanocytes make dark or light pigment in reponse to paracrine signals from the underlying dermal papilla.
In fact, there is a rare type of serval that has smaller spots than normal, a pattern referred to as "servaline", and that was originally thought to represent a separate species (10). While we have not yet obtained DNA from a servaline serval, a potentially homologous situation already exists in the Abyssinian and related breeds of domestic cats. A hallmark of the Abyssinian breed is the absence of dark tabby markings, showcasing how individual hairs are banded with different colors, a phenotype referred to as "Ticked". Chris and Kelly discovered that all Ticked cats carry loss-of-function mutations in Dkk4, and that loss-of-function for Dkk4 alters the molecular pre-pattern of gene expression in fetal skin. The effects on color pattern are beautifully apparent in a family of Savannah kittens: heterozygosity for a Dkk4 mutation causes the number of dark spots to increase and shrink in size, recapitulating the exact difference between a normal serval and a servaline serval.
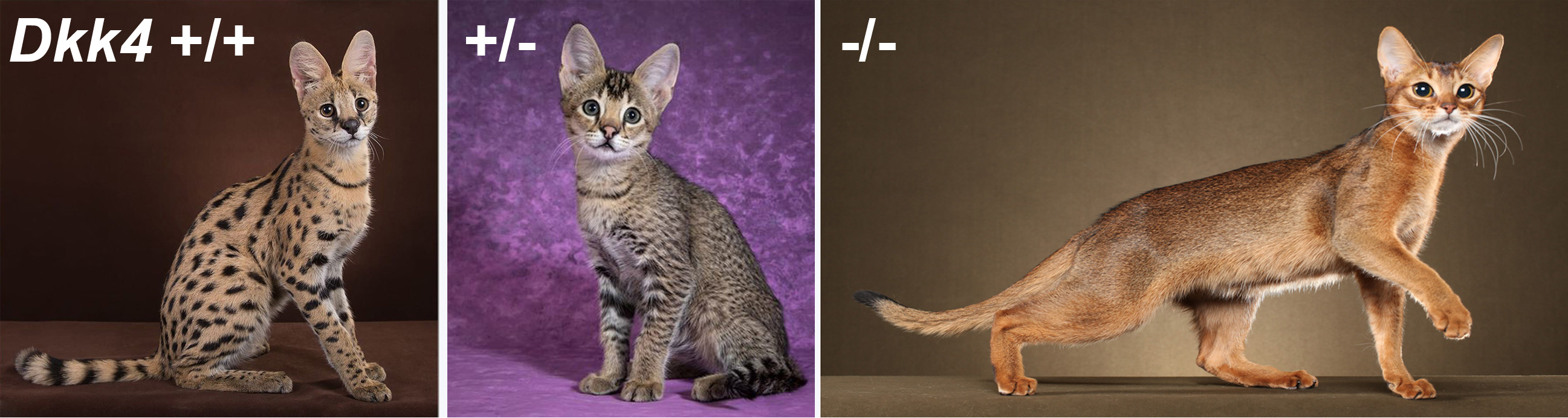
Figure 4. Genetic variation in spot number and size. Compared to the normal serval phenotype (left), heterozygosity for a Dkk4 mutation (middle) in a servaline Savannah cat causes dark tabby markings to become smaller and more numerous. Homozygosity for a Dkk4 mutation in an Abyssinian cat (right) further reduces the size of markings so that they are not easily visualized. Photos are by Jamila Agaeva (elft, middle) and Helmi Flick (right).
For lovers of cats and Rudyard Kipling, our work leads us to think differently about the Ticked phenotype in Abyssinian cats—instead of the absence of dark tabby markings, we propose that the markings have increased in number and decreased in size to the extent that they're just not readily apparent. For scientists, our work illuminates critical components of a self-organizing molecular process first conceptualized by Alan Turing, that like the signaling pathways controlling hair color, is likely to participate in aspects of biology beyond pigmentation. Looking forward, we wonder what and how variation in other genes affects the molecular pre-pattern, how the cell fate decisions signified by the molecular pre-pattern are maintained and implemented through skin development, and how changes in the pre-pattern explain cheetah spots, leopard rosettes, and tiger stripes.
References
- Bennett DC, Lamoreux ML. The color loci of mice--a genetic century. Pigment Cell Res. 2003; 16(4) 333-344. PubMed PMID: 12859616.
- Steingrímsson E, Copeland NG, Jenkins NA. Mouse coat color mutations: from fancy mice to functional genomics. Dev Dyn. 2006; 235(9) 2401-2411. PubMed PMID: 16691561.
- Kaelin CB, Xu X, Hong LZ et al. Specifying and sustaining pigmentation patterns in domestic and wild cats. Science (New York, NY). 2012; 337(6101) 1536-1541.
- Kondo S, Miura T. Reaction-diffusion model as a framework for understanding biological pattern formation. Science. 2010; 329(5999) 1616-1620. PubMed PMID: 20929839.
- Murray JD, Oster GF. Generation of biological pattern and form. IMA J Math Appl Med Biol. 1984; 1 51-75. doi:10.1093/imammb/1.1.51. PubMed PMID: 6600092.
- Turing AM. The chemical basis of morphogenesis. Philosophical Transactions of the Royal Society of London, Series B. 1952; 237 37-72. doi:10.1007/BF02459572.
- Meinhardt H, Gierer A. Pattern formation by local self-activation and lateral inhibition. Bioessays. 2000; 22(8) 753-760. PubMed PMID: 10918306.
- Chien AJ, Conrad WH, Moon RT. A Wnt survival guide: from flies to human disease. J Invest Dermatol. 2009; 129(7) 1614-1627. PubMed PMID: 19177135. PMCID: PMC3125088.
- Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006; 25(57) 7469-7481. PubMed PMID: 17143291.
- Ogilby W. Felis servalina. Proceedings of the Zoological Society of London. 1839; 7 94.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in