How to handle cellular DNA? Chromosomes and plasmids as substrates for condensins.
Published in Microbiology

Bacteria show a remarkable degree of cellular organization and many principles of cellular self-organization are highly conserved from bacteria to man. One of the key problems all cells need to cope with is the condensation of their DNA content and the faithful replication and segregation of the genetic material to daughter cells. Proteins involved in chromosome dynamics have therefore been studied for a long time and especially condensin complexes being ubiquitous in all domains of life have raised significant interest in recent years. Condensins are a conserved protein machinery that enables DNA compaction. Interestingly, bacterial condensins come in two flavors. Classical SMC-ScpAB (SMC is the abbreviation of “Structural maintenance of chromosome”) systems are found in Bacillus, Caulobacter and most other bacteria. In gamma proteobacteria such as E. coli a distantly related MukBEF system is present. However, both condensin types act on chromosome compaction and are therefore functionally equivalent. However, the situation in non-model species remained unclear.
Our lab has been working on the Gram-positive actinobacterium Corynebacterium glutamicum for several years now. As other actinobacteria, it grows by apical cell wall synthesis and has a rod-shaped morphology. We got interested in this species, when we realized that cell elongation and cell division are differently regulated compared to well-studied rod-shaped bacteria. The lack of knowledge on how C. glutamicum grows and divides was surprising, since industry uses this species since the 1960s to produce amino acids as food or feed additives on a large scale. Furthermore, closely related strains such as C. diphteriae and Mycobacterium tuberculosis are important pathogens and knowing molecular details of DNA-replication, growth and division is pivotal to identify potential molecular target points for antimicrobial agents. Early experiments in our lab suggested that organization of the corynebacterial chromosome plays a vital role for growth and even morphogenesis. However, little was known about the cell cycle in C. glutamicum. Kati Böhm than started to unravel the cell cycle and she soon realized that C. glutamicum contains at least two chromosomes stably tethered to the cell pole area and hence, these bacteria are indeed diploid. Therefore, chromosome segregation and compaction may very well be quite different from other rod-shaped bacteria. A bioinformatics search revealed that C. glutamicum (and its relatives) encode genes for two condensin systems, a SMC-ScpAB and a Muk-related MksBEFG complex. In order to study the functions of these systems we teamed up with Drs. Martial Marbouty and Romain Koszul from the Institute Pasteur in Paris.
Using genetic and cell biology approaches we were able to show that SMC is loaded onto the chromosome at specific DNA-sequences termed parS sites in a ParB-dependent manner, just like in other organisms such as B. subtilis and C. crescentus. In addition, chromosome conformation capture experiments (Hi-C) revealed that SMC complexes act on chromosome folding by juxta positioning the two replichore arms. Hi-C data further provided clear evidence that the origin region of the chromosome is segregated over the existing nucleoid towards midcell in a ParB-dependent manner. This segregation is an exquisite example of cellular self-organization and reveals a structural role for the chromosome beyond encoding genetic information.
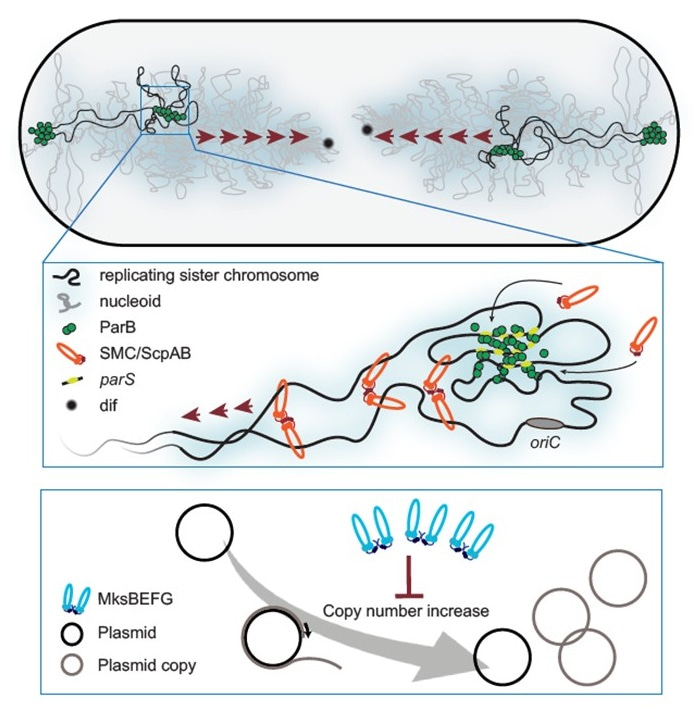
To our surprise, however, the alternative condensin system MksBEFG had no effect on chromosome conformation. Instead, we found that Mks proteins localize to the cell pole and interact with the polar scaffold protein DivIVA. Instead, we found that Mks proteins localize to the cell pole, where they interact with the polar scaffold protein DivIVA, and that cells expressing a fluorescent fusion of MksB show an increase in polar fluorescence when cells were transformed with a plasmid. This immediately raised the idea that MksB could react to the presence of plasmid DNA. This idea was supported by a very recent finding from the Sorek lab in Israel that suggests that MksBEFG systems could be a type of antiviral or anti-plasmid defense system. Therefore, we analyzed plasmid maintenance in strains lacking MksB. Our data revealed that high copy number plasmids were not affected by Mks, but low or single copy plasmids were enriched up to 60 fold in absence of MksB. Thus, Mks-like condensins have evolved to restrict plasmid copy number and are so far MksBEFG is the only known condensin systems amongst pro- and eukaryotes that is not involved in chromosome organization.
The published article can be found in Nature Communications here: https://rdcu.be/b3qmz
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in