Hunting for the authentic host of giant viruses in the marine environment
Published in Microbiology

Marine viruses, estimated to outnumber the stars in the universe, infect and kill billions of marine microbes every day. This drives nutrient cycling across marine ecosystems, thus influencing global climate and the marine food web. The discovery of giant viruses has revolutionized our understanding of viral life cycles and ecological impact. Giant viruses inhabit ecosystems all over the globe and can infect many clades of protists, which is a general term for eukaryotic microbes. Some giant viruses infect and kill bloom-forming algae, playing an essential role in the marine ecosystem.
In light of their ecological importance, extensive efforts have been made to catalog the vast diversity of giant viruses by using metagenomic studies conducted at the bulk population level. On the other hand, the life cycle of giant viruses is primarily studied under controlled lab conditions using only limited host model systems. These model systems may not necessarily be the viruses’ native host in nature. We therefore pose the question: “How can we identify the authentic host of giant viruses in nature?”. A better understanding of giant virus infection dynamics in the natural ecosystem will highlight their ecological importance.
New single-cell RNAseq (scRNA-seq) technologies bring new opportunities for exploring host-virus interactions on unprecedented resolution.
Our study, which results from a fruitful collaboration between the Vardi and Aylward labs, tried to bridge this knowledge gap by studying the natural protist population infected by giant viruses one cell at a time. Our collaboration combined the experience of the Vardi Lab with single-cell transcriptomics and protist-virus interactions and the Aylward Lab’s expertise in giant virus genomics and phylogeny. We applied a novel scRNA-seq method used extensively in the animal and plant world to map and characterize cell types during development and in immune systems.
We used this approach to analyze samples from the natural marine environment and to capture transcriptional profiles of both the protist host and giant viruses that infect them among thousands of individual cells. This analysis identified previously unknown host-virus interactions in the marine environment.
Elucidating the host-virus network at the resolution of a single-cell
In May 2018, we conducted a field experiment in Raunefjorden near Bergen, Norway, where we induced an Emiliania huxleyi algal bloom by adding nutrients to natural plankton populations and followed the bloom for 24 days. We then sequenced the samples we collected using the scRNA-seq technique. In this method, each cell gets a unique barcode that enables us to quantify each RNA molecule in each cell. We used the conserved 18S ribosomal RNA gene to identify the host cells and conserved giant virus gene markers to identify the actively infecting viruses.

The setting of the field experiment in Bergen, Norway.
a, The experiment included seven bags submerged in the fjord and filled with fjord water to capture natural plankton communities. b, An algal bloom was induced by adding nutrients to the water in the bags. The milky shade of the water in the bag indicates the high concentration of the algae Emiliania huxleyi.
Using this method, we analyzed tens of thousands of cells and revealed hundreds of host cells actively infected by diverse giant viruses. We found many new protist-host pairs; some detected hosts belonged to groups for which there was no known virus, and some detected viruses had no known host. Thus, we show that this method can illuminate host-virus interactions that have yet to be studied.
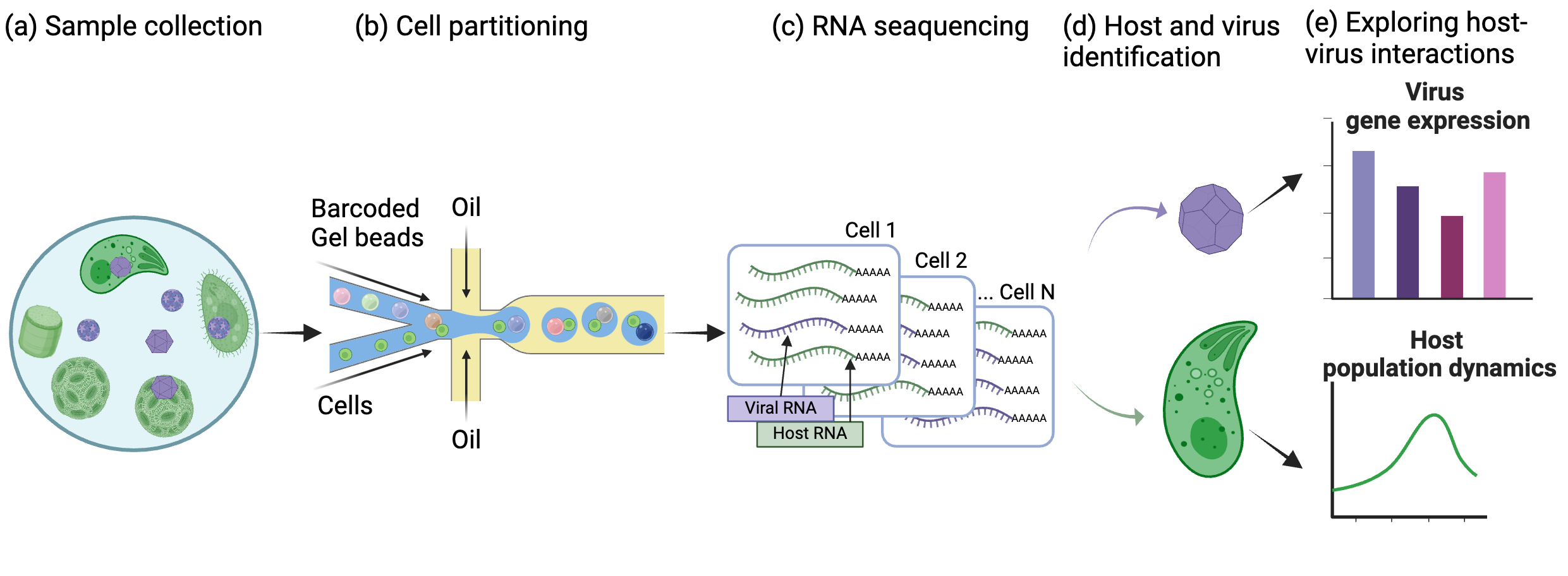
An overview of the pipeline for detecting host-virus pairs in the natural environment.
a, Samples were collected from the natural environment. b, Resuspended cells were partitioned by a 10X Chromium microfluidic device. Partitioned cells were combined with beads containing cell-specific barcodes and sample barcodes. c, RNA from each cell was sequenced. d, Host and virus were identified from each cell using sequence homology. e, After identifying host and virus pairs, we further analyze the genes expressed by the virus and its impact on the host population.
We then used this newly acquired information to draw a picture of active viral infections co-occurring in diverse protist populations within the algal bloom. The abundance of one dominant species, in our case, E. huxleyi, often masks the less abundant species when studying the bulk population. By looking at the level of single cells, we could better capture the rare, infected cells that can contribute to the population dynamics and ecology.
One such rare subpopulation belonged to a protist from class Katablepharidaceae, making up less than 0.5% of the identified cells. Very little is known about this protist, and it has no known virus. We identified the virus that infects this rare protist and correlated the appearance of the virus to the demise of its population.
The fact that these organisms are rare does not necessarily mean that they have a minor role in the ecosystem; on the contrary- they might have a more prominent role than we think. Studies have highlighted the importance of rare species as having a disproportional impact on the ecosystem. Rare species are often more active than abundant species, have a high per-organism contribution to community activities, and enhance the functionality of abundant species. Moreover, active infection by rare viruses can serve as a seed bank population for subsequent infections. Recent studies on viral infection in the natural environment have shown that low infection levels are widespread in marine ecosystems and may have significant consequences for viral persistence over broad geographic areas.
Overall, this approach will help draw a holistic picture of natural single-cell populations and the viruses that infect them in the marine environment. This approach can be adapted and expanded to other viruses, such as RNA viruses, and other ecosystems, such as soil and freshwater.
We thank all the contributors to this project: Prof. Frank Aylward, Dr. Gur Hevroni, Dr. Daniella Schatz, Dr. Flora Vincent, and Dr. Carolina A. Martinez-Gutierrez.
References
- Schulz F. et al. Giant virus diversity and host interactions through global metagenomics. Nature 578, 432–436 (2020).
- Endo, H. et al. Biogeography of marine giant viruses reveals their interplay with eukaryotes and ecological functions. Nature Ecology & Evolution 4, 1639–1649 (2020).
- Aylward, F. O., Moniruzzaman, M., Ha, A. D. & Koonin, E. V. A phylogenomic framework for charting the diversity and evolution of giant viruses. PLoS Biol 19, e3001430 (2021).
- Moniruzzaman, M., Martinez-Gutierrez, C. A., Weinheimer, A. R. & Aylward, F. O. Dynamic genome evolution and complex virocell metabolism of globally-distributed giant viruses. Commun. 11, 1710 (2020).
- Ku, C. et al. A single-cell view on alga-virus interactions reveals sequential transcriptional programs and infection states. Adv.6, eaba4137 (2020).
- Hevroni, G., Vincent, F., Ku, C., Sheyn, U., & Vardi, A. Daily turnover of active giant virus infection during algal blooms revealed by single-cell transcriptomics. Science Advances. 9, 41 (2023)
- Vincent, F. et al. Viral infection switches the balance between bacterial and eukaryotic recyclers of organic matter during coccolithophore blooms. Commun. 14, 1–17 (2023).
- Jousset, A. et al. Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J. 11, 853–862 (2017).
- Breitbart, M. & Rohwer, F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13, 278–284 (2005).
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in