β-Hydroxylation of α-amino-β-hydroxylbutanoyl-glycyluridine catalyzed by a nonheme hydroxylase ensures the maturation of caprazamycin
Published in Chemistry

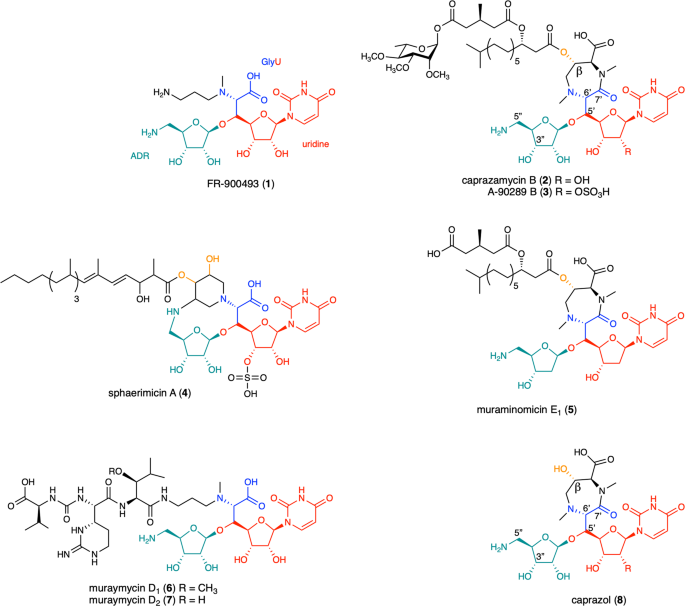
Caprazamycin is a nucleoside antibiotic that inhibits phospho-N-acetylmuramyl-pentapeptide translocase (MraY). The biosynthesis of nucleoside antibiotics has been studied but is still far from completion. The present study characterized enzymes Cpz10, Cpz15, Cpz27, Mur17, Mur23 out of caprazamycin/muraymycin biosynthetic gene cluster, particularly the nonheme αKG-dependent enzyme Cpz10. Cpz15 is a β-hydroxylase converting uridine mono-phosphate to uridine 5′ aldehyde, then incorporating with threonine by Mur17 (Cpz14) to form 5′-C-glycyluridine. Cpz10 hydroxylates synthetic 11 to 12 in vitro. Major product 13 derived from mutant Δcpz10 is phosphorylated by Cpz27. β-Hydroxylation of 11 by Cpz10 permits the maturation of caprazamycin, but decarboxylation of 11 by Mur23 oriented to muraymycin formation. Cpz10 recruits two iron atoms to activate dioxygen with regio-/stereo-specificity and commit electron/charge transfer, respectively. The chemo-physical interrogations should greatly advance our understanding of caprazamycin biosynthesis, which is conducive to pathway/protein engineering for developing more effective nucleoside antibiotics.
Follow the Topic
-
Communications Chemistry

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the chemical sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
f-block chemistry
Publishing Model: Open Access
Deadline: Feb 28, 2026
Experimental and computational methodology in structural biology
Publishing Model: Open Access
Deadline: Apr 30, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in