Immunophenotyping COVID-19 patients during the first epidemic wave in Singapore
Published in Microbiology

SARS-CoV-2, the virus responsible for COVID-19 disease, first appeared at the end of 2019 in China. At the start of January 2020, my boss, Prof Lisa Ng, had hoped that the outbreak would not spread, but sounded the alarm in the lab and galvanized us to look at the reported viral sequences that were coming out from China as a precautionary measure. With foresight, she alerted us to start preparing for a potential epidemic and to think about the contributions that we could make to help support the clinicians in Singapore and the world and start stocking up reagents before the provider circuits would be down. At the end of March 2020, when the virus was spreading rapidly in Singapore, Lisa, after jumping many administrative hoops, finally obtained the green light to start immunophenotyping the blood of COVID-19 patients. From the preprint literature (not yet peer reviewed) that we were seeing, we suspected that many immune cell types would show drastic changes during the course of infection and they needed to be characterized properly (Figure 1). Understanding these would allow two things. Discover early cellular markers that could associate with severity and possibly predict it for patient triage. And understand the immune imbalances during SARS-CoV-2 infection to identify targets for immune modulation therapies.
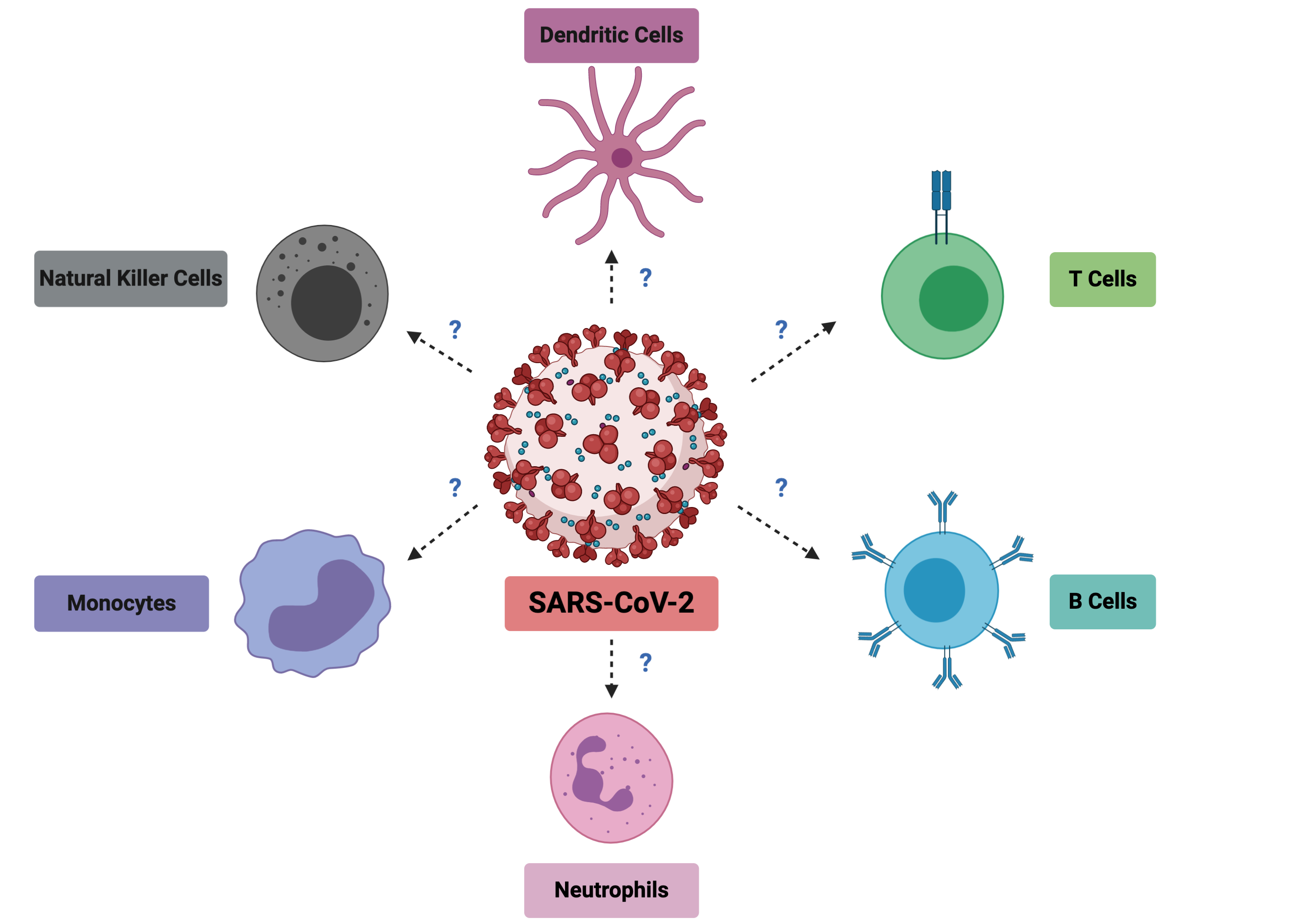
As SARS-CoV-2 continued to rampage the world, resulting in the loss of lives and livelihoods, this project needed to be done at lightning speed. At the Singapore Immunology Network where I work in A*STAR, we combined the expertise of a talented T-cell and flow cytometry expert (Dr. Weili Xu), not less talented neutrophil expert (Dr. Immanuel Kwok) and myself (a virologist). Together, with the help of a few colleagues who were allowed to work during Singapore’s “Circuit Breaker”, we immunophenotyped 54 acute patient samples, healthy donors and samples from COVID-19 recovered patients with three flow cytometry panels designed to partially overlap but still assess as many immune cell types as possible. After this was completed came the interesting part, the analysis! We analyzed, compiled the different immune cell counts and their phenotypic and activation markers. Subsequently, our clinical partners provided us with the patients’ clinical data which allowed separating patients into four different severity groups. From there, we investigated all the immune cell populations changes that were associated with severity.

Out of all the subsets we identified, the most interesting were the immature neutrophils and gamma delta 2 T cells (VD2 cells). These particular immature neutrophils increased in spectacular fashion in the blood of patients with high disease severity, while VD2 T cells showed a granular decrease which followed severity with a low standard deviation. We then looked at these cell numbers in patient samples at early time points after diagnosis. That was when we realized that we had found something that could prove useful in the clinical setting to potentially predict if patients will develop severe symptoms of pneumonia or hypoxia, and decide to preventively admit and/or treat them accordingly (Figure 2).
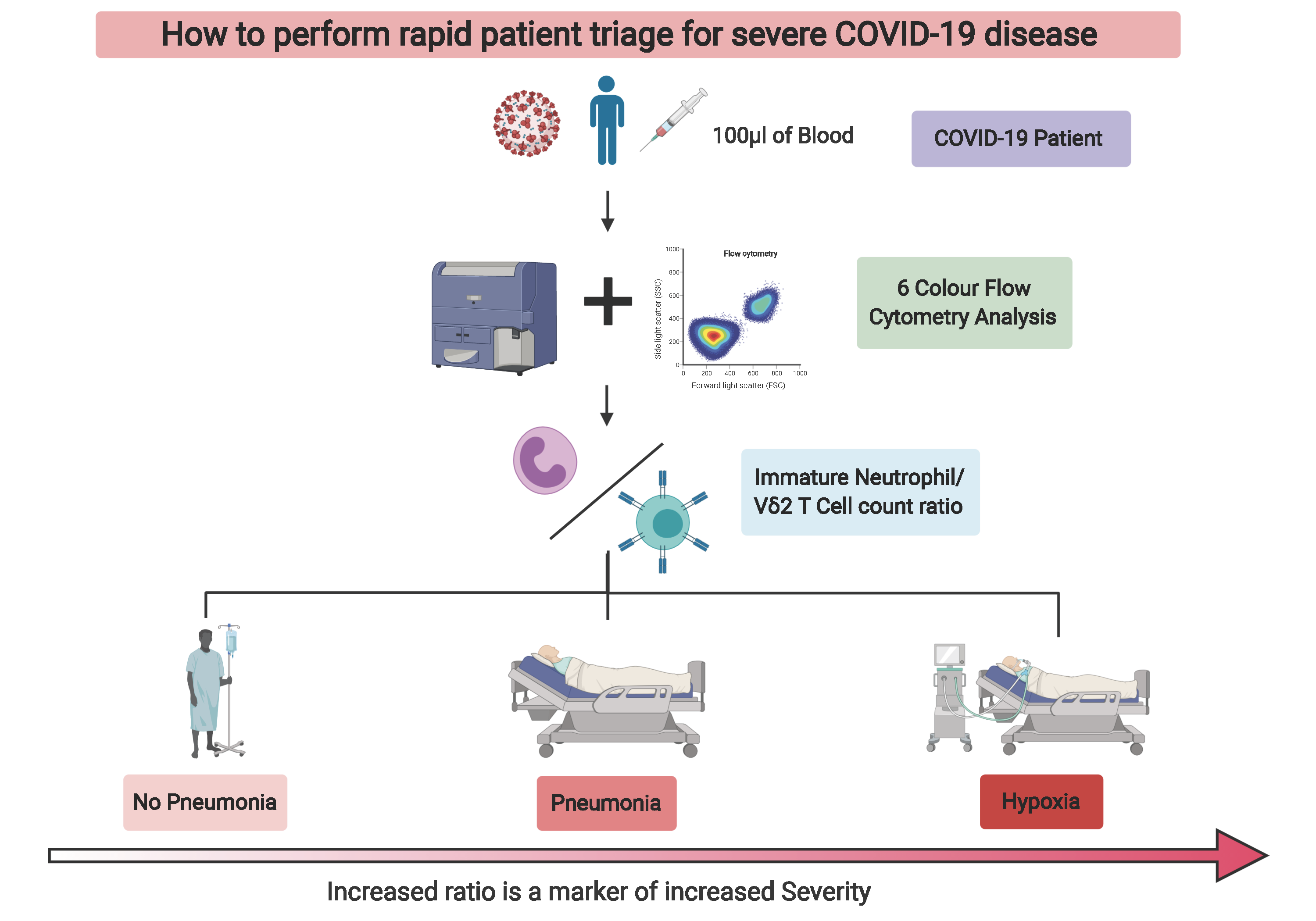
We discovered that with a 6-color flow cytometry compatible with most standard cytometers in diagnostic laboratories, one could potentially facilitate patient triage and significantly ease the healthcare burden in times of crisis. Now, all that was left was to convince our peers: the reviewers, as you will be able to read in the transparent review process that accompanies our manuscript. That took three stressful and challenging months, longer that what it took to make the discovery (preprint was available 12th of June 2020). Nonetheless, we believe that this is still in time to help healthcare systems cope with SARS-CoV-2 infections while waiting for much needed efficient vaccines.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in