Improved high-quality reference genome of red drum facilitates the resistance-related gene exploration
Published in Genetics & Genomics
The Red drum (Sciaenops ocellatus) was originally from the western Atlantic Ocean from Massachusetts of the United States to the northern coast of Mexico, and has been introduced to 11 countries and regions around the globe, becoming a world-wide cultured fish species. It characterized by fast growth rate (world record ~40kg; Chinese record ~26kg), miscellaneous diet, strong reproduction ability, resistance to disease, tolerance to low oxygen (≥2.2mg/L), etc., and could adapt to diversified habitats quickly. Since its introduction to the mainland of China in 1991, the cumulative aquaculture production of S. ocellatus has reached 955,159 tons, accounting for 94% of the world's total aquaculture production. Among them, cage culture accounted for more than 90% of the production. However, escapes due to extreme weather (e.g., typhoons) have caused great pressure on local ecosystems and biodiversity.
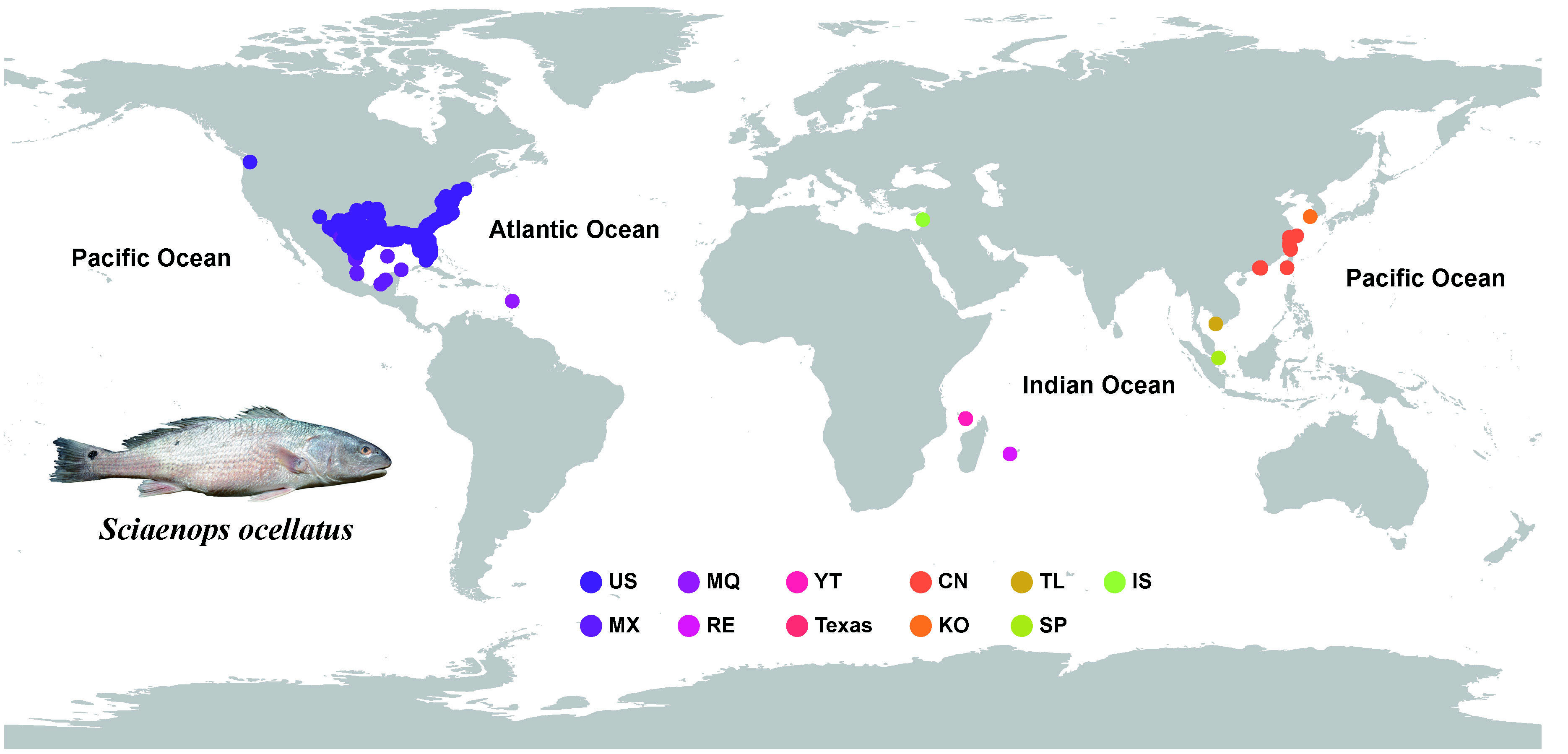
US: United States of America, MX: Mexico, MQ: Martinique, RE: La Réunion, YT: Mayotte, CN: China, KO: Korea, TL: Thailand, SP: Singaporean, IS: Israeli.
The research team of Jing Liu and Yongshuang Xiao at the Institute of Oceanology Chinese Academy of Sciences (IOCAS) completed a study on the early warning ecology of the invasion of S. ocellatus, an exotic aquaculture species. It was found that, in addition to the Gulf Coast, S. ocellatus was still present in three hotspots around the world (Uruguayan coast, Chinese coast, and Australian coast), and that, with the global warming, the center of mass of its distribution area would be changed by the global warming. As the global climate warms, the center of mass of S. ocellatus will shift northward and expand significantly (448,178.6 km2), and Bohai Bay (an important spawning ground for fishery organisms) will become a new hotspot for S. ocellatus. Stable isotope analyses showed that the trophic level of the escaped S. ocellatus was significantly higher than that of typical indigenous fishes in the East China Sea (e.g., fishes of the family Sciaeninae), and was in the same level with other aggressive predatory fishes (e.g., Trichiurus lepturus, Lophius litulon, and Anago anago), which were highly ecologically competitive. The research results provided an ecological basis for the evaluation of the invasion potential of the exotic species S. ocellatus.

On the basis of this study, the research team of Jing Liu and Yongshuang Xiao constructed a high-quality genome of S. ocellatus at the chromosome level by using PacBio CCS and Hi-C technologies, and obtained a genome size of 725.33 Mb and a Contig N50 of 28.30 Mb. The adaptation and regulatory mechanisms of S. ocellatus in extreme environments were systematically analyzed and evaluated from the molecular genetic perspective by using low-salt-gradient stress experiments, Gene Set Enrichment Analysis (GSEA), and Weighted Gene Co-Expression Network Analysis (WGCNA) methods. The research team successfully identified 54 core genes that were closely related to environmental stresses, and these genes played an important role in the adaptation of S. ocellatus to low-salt environments. For the first time, it was found and demonstrated that S. ocellatus stimulates endoplasmic reticulum stress response in gill tissues after entering into the hypolimnetic environment; in order to adapt to hypolimnetic stress, S. ocellatus adapted to the hypolimnetic environment by rapidly activating four ion transport channels (Integrin β sensor: cxcr4, Itgb8; TRPV sensor: trpv4/6, aqp3; NCC sensor: prlr, slc12a3; NHE3 sensor: slc9a3, rhcg1) to rapidly adapt to the hypotonic environment and maintain cellular homeostasis by maximizing high levels of calcium ions in the endoplasmic reticulum (endoplasmic reticulum uptake of calcium ions in the cytoplasm). Meanwhile, S. ocellatus suppressed the mitochondrial expression by decreasing Vdac1/2 expression Cytochrome C expression, which in turn inhibited apoptosis. These findings provided a molecular genetic basis for the evaluation of the invasion potential of the alien species S. ocellatus, and a theoretical basis for the development of its invasion prevention and control countermeasures.
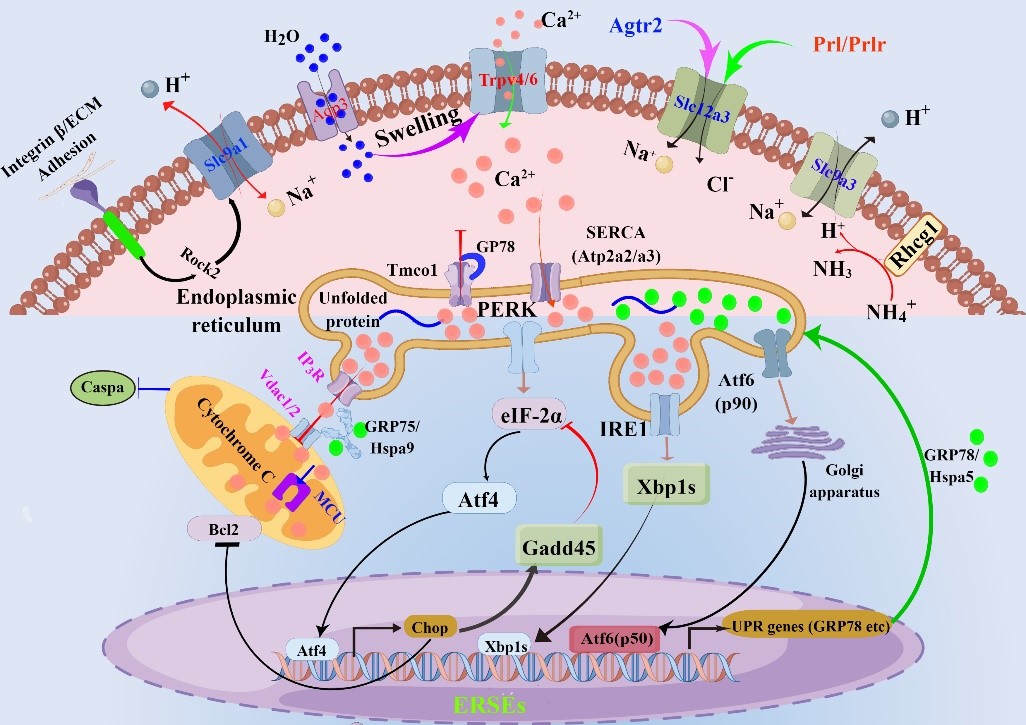
Rapid regulation process of hypo-osmotic ecological adaptation in the S. ocellatus
We are working with collaborators to expand the S. ocellatus invasion assessment to countries and regions of key introductions such as South Africa, Uruguay. Our main research directions remain the evaluation of the environmental adaptive capacity of S. ocellatus and the assessment of genetic adaptation and differentiation in transplantation sites. On the basis of sufficient scientific evidence, we will draw attention and concern, brainstorming to propose S. ocellatus quality invasion prevention and control strategies, and provide early warning and prevention and control strategies to prevent S. ocellatus from global invasion events.
Follow the Topic
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
Publishing Model: Open Access
Deadline: Apr 17, 2026
Data to support drug discovery
Publishing Model: Open Access
Deadline: Apr 22, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in