In vitro construction of the COQ metabolon unveils the molecular determinants of coenzyme Q biosynthesis
Published in Chemistry and Cell & Molecular Biology

Introduction
Coenzyme Q, CoQ, or more commonly, ubiquinone, is renowned for its function as an electron shuttle and conduit in the electron respiratory chain. CoQ also governs a whole host of different reactions and is tied to many metabolic pathways including fatty acid oxidation, mitochondrial uridine biosynthesis and more recently, of mounting interest, ferroptosis. CoQ is considered one of the most hydrophobic molecules known in nature and is synthesised in the mitochondria at the interface between the inner mitochondrial membrane and the matrix and comprises two juxtaposing ends: a highly hydrophobic poly-isoprenoid tail that anchors the antioxidant within phospholipid bilayers and a fully substituted aromatic head group responsible for its redox properties (Fig. 1a).
In animals, biosynthesis of CoQ is currently attributed to ten different proteins with COQ3, COQ4, COQ5, COQ6, COQ7, and COQ9 forming the iconic COQ metabolon. Metabolons are protein assemblies that perform a series of reactions in a metabolic pathway (Fig. 1b). They have been shown to channel intermediates of the reaction coordinate and assemble upon given cellular needs. However, many questions regarding the aptitude of metabolons for enzyme catalysis and their dynamic behaviour remains poorly understood. Here, the COQ metabolon, a composite of COQ proteins, adheres to the matrix side of the inner mitochondrial membrane to build the final aromatic ring of CoQ. These include COQ3, COQ4, COQ5, COQ6, COQ7 and COQ9. The formation and regulation of the COQ metabolon is postulated to be facilitated by COQ8, which in humans is present as two paralogs, COQ8A and COQ8B.
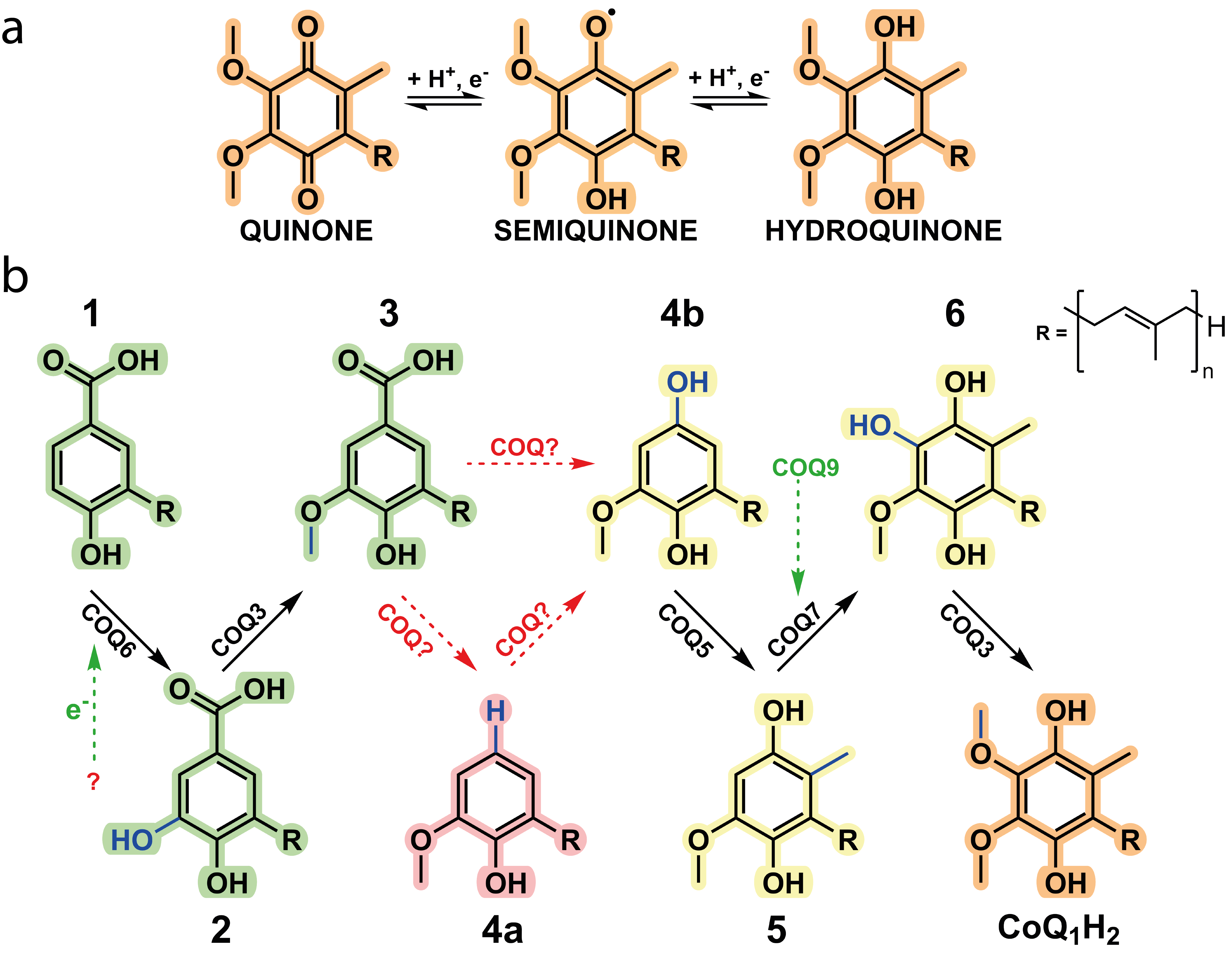
Figure 1. Coenzyme Q is a redox mediator.
Why the COQ Metabolon?
The functional roles behind several of the animal COQ proteins remains bereft in our biochemical understanding. Indeed, the molecular determinants essential to C1 modification of the CoQ precursor and physiological roles of COQ8 in CoQ biosynthesis are unclear. Further questions include: is the formation of the COQ metabolon, or subcomplexes, essential for enzymatic activity and does further protein complexation bolster enzymatic activity? Additionally, how is CoQ biosynthesis regulated? Major challenges associated to these questions are due to: (i) the extreme hydrophobicity and instability of the intermediates of the biosynthetic pathway; (ii) the poor stability of the COQ partners as stand-alone systems. As such, building and investigating metabolons, in general, in vitro is arduous.
Methodological Strategy
To address these questions and uncover the roles of the COQ proteins at the helm of animal CoQ biosynthesis within the COQ metabolon, herein this article, we employ an innovative evolution-engineering approach to build the COQ metabolon in vitro. We integrate ancestral sequence reconstruction, ASR, as a methodological backbone to our biochemical study as a means to build experimentally tractable systems with heightened stability. To overcome issues with substrate solubility, we turn to CoQ precursors possessing a single isoprenoid unit. Our approach, based on the reconstruction of the full-scale biosynthetic pathway, unveils all the enzymatic steps in coenzyme Q biosynthesis and demonstrates the functional coupling among the enzymes forming the metabolon.
Major findings
We were able to isolate and purify each of the COQ proteins pertaining to the metabolon and characterise their role within the biosynthetic pathway. As previously speculated, COQ6 represents the initiator and hydroxylates the C5 position of the aromatic headgroup. More importantly, this work illustrates that a ferredoxin coupled pair, Ferredoxin Reductase (FDXR) and Ferredoxin 2 (FDX2), are required for COQ6 reduction and activity. The previously documented functions of COQ3, COQ5 and COQ7 were corroborated in this research and their kinetic parameters were measured. Of great interest, however, concerns the enzymes responsible for the modification at the C1 position which have long remained enigmatic.
Here in this research, using a combination of both analytical and spectrophotometric techniques, we observed that both COQ4 and COQ6 were responsible for the C1-transformation. COQ4 was identified to be a Zn2+-dependent aromatic decarboxylase, producing the non-substituted C-H group at the C1 position. These results suggested that instead of performing an oxidative decarboxylation reaction, a non-oxidative decarboxylation reaction is performed. Subsequently, COQ6 was observed to turnover the formed product, performing a hydroxylation at the C1 position, a reaction chemically equivalent to its previous C5-hydroxylation. This result illustrates that COQ6, similarly to COQ3, possesses dual functionality. Strikingly, pooling together COQs 3, 4, 5, 6, 7 and 9, resulted in the accumulation of Coenzyme Q, demonstrating that these enzymes are responsible for its biosynthesis. Removing each enzyme in turn resulted in the accumulation of its given substrate, with no other downstream enzyme reactions taking place. This confirmed the dual functionality of COQ6 and COQ3, the role of COQ4 as the decarboxylase and that the CoQ biosynthetic pathway possesses a firm directionality. Of note, however, in the presence of COQ8 the yield of CoQ increased approximately five times and the presence of intermediates disappeared entirely. These results highlight the importance of COQ8 in substrate turnover and illustrate the formation of the metabolon as molecular channelling is evident. The specific molecular mechanisms underpinning this result will be of great interest and scrutiny for future research.
Outlook
In this work, to overcome hurdles associated to both COQ and CoQ intermediate stability, we employed ASR and took advantage of more soluble mono-prenylated CoQ intermediates. Remarkably, this evolution-engineering approach successfully generated enzymatically competent enzymes, in line with current literature, and conveyed key characteristics that support metabolon generation. More importantly, however, this strategy was able to characterise the unknown reaction steps in CoQ biosynthesis and assign the enzymes governing these transformations in animals (Fig. 2). We do not see any major reason as to why this methodology could not be translated to other metabolons more generally and we implore its application to resolve enzymatic behaviour for onerous and challenging protein studies.
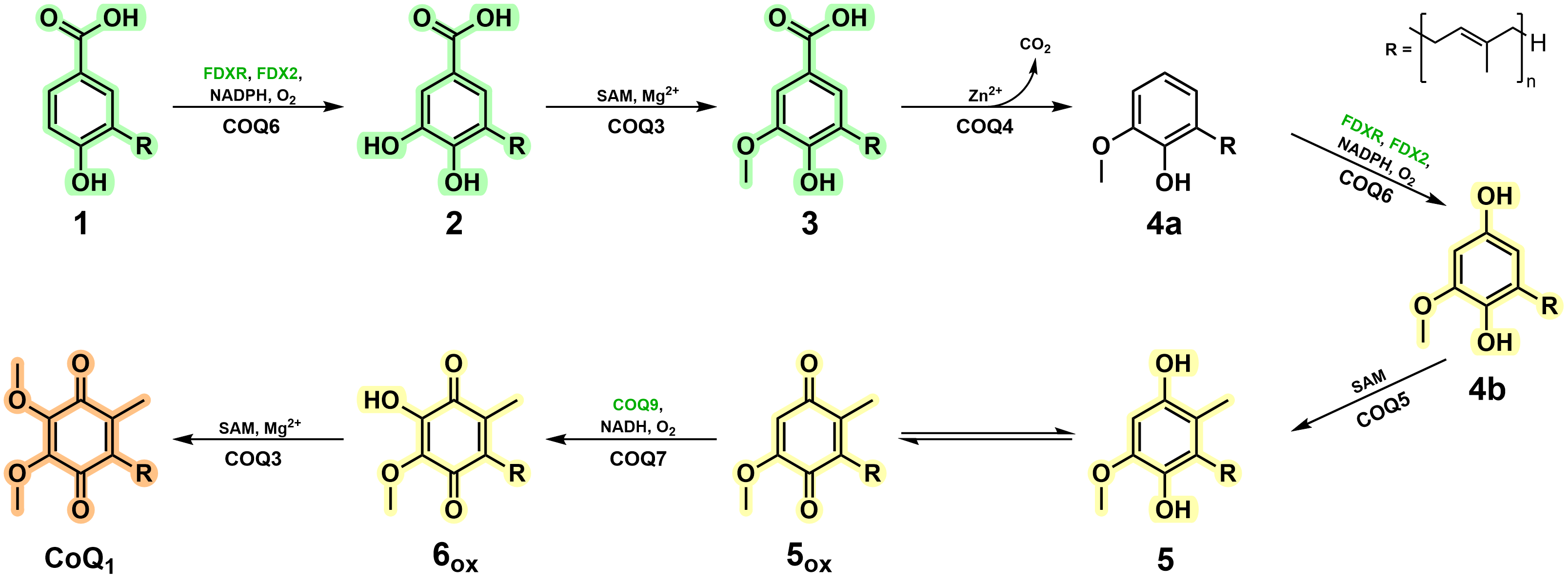
Figure 2. Vertebrate CoQ biosynthetic pathway in light of the findings of this work.
Large protein complexes typically suffer, or fail altogether, when proteins are stripped from the core. Our results portray that, instead, in terms of enzymatic function, the systems comprising metabolons are not so fragile per se; the individual protein components are functional as stand-alone systems yet lack a sense of co-operativity when simply mixed. Alternatively, in the presence of their key chaperoning/organising partners, in this case COQ8, enzymatic activity is propelled and streamlined as illustrated by the lack of intermediate accumulation and increased CoQ yields. Hence, COQ8 captures the true power and essence of a metabolon - transitioning independently functional enzymes into a cohesive biosynthetic macromolecular machine.
Future questions
With the metabolon assembled and experimentally tractable, we now possess an idyllic molecular setting for further biochemical investigation: (i) how do metabolons function and traffic substrates/intermediates; (ii) what is the molecular composition of the COQ metabolon and do metabolons, more generally, possess a defined structure; (iii) can metabolons represent larger drug targets that can be attacked to thwart a metabolic pathway all together; (iv) how is COQ8 facilitating biocatalysis and more generally, do other metabolons possess supervisory and managerial proteins that organise the macromolecule for catalysis?
Follow the Topic
-
Nature Catalysis

This journal brings together researchers from across all chemistry and related fields, publishing work on homogeneous catalysis, heterogeneous catalysis, and biocatalysts, incorporating both fundamental and applied studies.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in