Inside the genome of mountaineer bird: Black Redstart and the secrets written in its DNA
Published in Ecology & Evolution and Genetics & Genomics
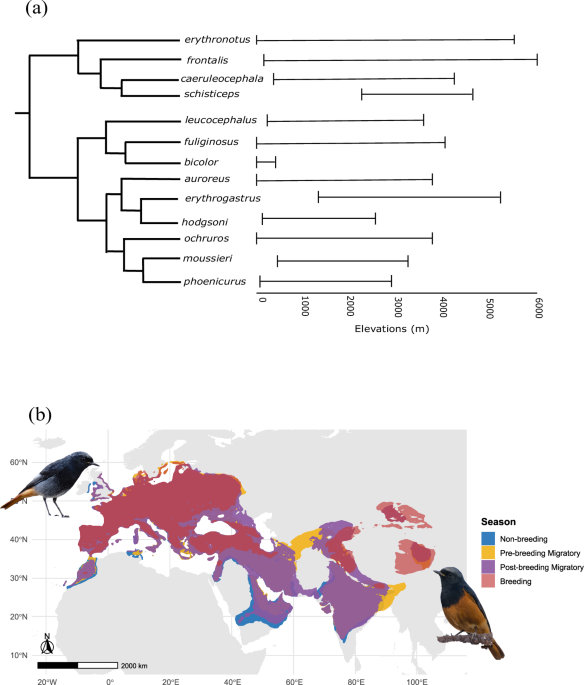
Birds have long fascinated humans - not just for their songs and colors, but for their incredible ability to adapt to diverse environments. Among these, the Black Redstart (Phoenicurus ochruros) is a remarkable survivor. Widely distributed across Europe, Asia, and North Africa, it thrives in habitats ranging from rocky cliffs and mountains to urban rooftops. Even more astonishing, it performs altitudinal migrations, moving from high-elevation breeding grounds above 4,000 meters to lower-elevation areas (0 - 2,500 meters) during the non-breeding winter season, just like a mountaineer but without oxygen cylinders, hiking gears and tents. This exposure to dramatically different environmental conditions - cold, snowy highlands versus milder lowlands - makes the Black Redstart an excellent model for studying physiological and genetic adaptation.
These questions inspired us to generate the first reference genome for the Black Redstart. A reference genome is a blueprint of an organism’s DNA, allowing us to explore genes, regulatory elements, and other genomic features that shape its biological adaptation. Basically, it’s like a detailed instruction manual telling us which genes exist and where to find them. These genes and other elements in their genome hold key to their survival. Until now, the Black Redstart and its group (Phoenicurus) lacked such a resource, limiting our ability to study its adaptations at the molecular level. Without a reference genome, studying DNA is like exploring a city with no map, not only you do not know where anything is, but you also can’t see the major roads, intersections, or neighborhoods.
Why the Black Redstart?
The Black Redstart’s wide geographic and altitudinal range makes it an ideal species for studying adaptation to challenging environments, particularly high-altitude habitats. It's relative abundance and tolerance of human-altered landscapes also makes sample collection feasible - a major consideration in genomic studies. Finally, as a passerine (a group that includes most songbirds), it provides opportunities for comparative genomics, helping us understand evolutionary patterns across related species.
Fieldwork at the edge of the Plateau
Our fieldwork took us to Qinghai Province in the Qinghai-Tibetan Plateau (QTP), a region known as “roof of the world” because its average elevation exceeds 4,000 meters (13,000 feet). We captured an adult female Black Redstart using mist-nets at an elevation of 4,400 meters.

Habitat of the Black Redstart at 3,000 m elevation in the Qinghai-Tibetan Plateau, Qinghai Province, China, along with members of our field team.
The bird lived in a small grassland town, where it was the only Redstart species present. Observing this species in its high-altitude habitat was fascinating experience. Black Redstarts nest in rocky ground or cliffs but recently they have been more creative and adapted to changing landscape by exploiting human structures, using gaps in buildings as nest sites. Even in August, the conditions were harsh: snow fall regularly, and nighttime temperatures drop below zero degree celsius. Interestingly, although the female we captured had completed her reproductive cycle, she had not yet migrated down to lower elevations.

These observations highlight the extreme environmental variability that Black Redstarts experience and underscore why understanding their genetic adaptations is so important. From surviving cold high-altitude nights with low oxygen conditions to navigating seasonal migrations, this small bird faces challenges that few other species encounter.
Challenges in sample collection and preservation
Collecting high-quality DNA from wild birds in such remote and extreme conditions presents significant logistical challenges. High-altitude fieldwork is physically demanding due to low oxygen levels, cold temperatures, and unpredictable weather. Handling birds under these conditions requires careful planning to ensure both researcher safety and minimal stress to the birds.
Once samples were collected, maintaining a strict cold chain is critical. DNA degrades rapidly if samples are exposed to heat or fluctuating temperatures, which can compromise DNA sequencing quality downstream. In the field, we immediately preserved tissue and blood samples in liquid nitrogen, then transported them under carefully controlled conditions back to the laboratory. This required coordination, reliable equipment, and contingency planning for power or transport interruptions in a remote, high-altitude environment.
Maintaining sample integrity was especially important for PacBio HiFi sequencing, which relies on ultra-high molecular weight DNA. Any degradation could reduce DNA sequence read length and accuracy, impacting the genome assembly. Despite these logistical hurdles, our careful field protocols allowed us to obtain DNA of exceptional quality, enabling the successful assembly of the first reference genome of Black Redstart.

Field team member holding an adult female Black Redstart for blood sampling. Samples were collected for high-quality DNA extraction while minimizing stress to the bird.
From Field to Genome
Once in the lab, we used PacBio HiFi long-read sequencing, a cutting-edge technology that generates long, highly accurate DNA sequence. Long reads are crucial because genomes aren’t simple texts - they contain repetitive sequences, duplicated regions, and “jumping genes” called transposable elements. These regions are difficult to assemble with older sequencing methods.
The final nuclear genome of the Black Redstart is 1.37 gigabases in size. It is assembled into 296 pieces (contigs), with half of the total genome contained in pieces that are 29.9 megabases or longer - a sign of a very continuous assembly.To ensure high quality assembly, we applied multiple validation methods. For e.g., BUSCO analysis confirmed that nearly all expected genes were present. Mapping the raw sequence reads back to the genome showed excellent coverage, and comparisons with other passerines indicated a biologically meaningful, high-quality assembly.
One unexpected finding was that a large portion of the Black Redstart’s genome (~ 30%) was made up of transposable elements. These are pieces of DNA that can copy or move around within the genome. This is the highest proportion reported for passerines second to previously reported genome of Bell’s sparrow. These transposable elements are known to influence gene regulation, offering clues about how Black Redstarts adapt to diverse and challenging environments.

The puzzle of Gene annotation
Sequencing the genome is only part of the story. To make it truly useful, we needed to identify genes and predict their function. For this, we combined three sources of evidence: RNA sequence data, which reveals genes that are functionally active; protein homology from related species; and computational predictions. Integrating these approaches allowed us to annotate 18,609 genes, providing a comprehensive map of the Black Redstart’s functional genome.
This process was not without challenges. RNA sequence data sometimes fails to align as expected, and some predicted genes initially don't match patterns observed in other birds. Each obstacle required troubleshooting, iterative testing, and careful validation. These moments were demanding but ultimately rewarding - each problem solved brought us closer to a genome we could trust.
Lessons from the lab and the field
One of the most important lessons from this project was the value of combining field biology with genomics. Knowledge of the species’ natural history - its altitudinal migration patterns, habitat preferences, and reproductive timing - guided our sampling and sequencing strategies. Conversely, the genome now provides a molecular lens to study the genetic basis of these ecological traits.
Patience and persistence were also key. Genome assembly and annotation are like solving an enormous, multidimensional puzzle: each piece must fit precisely, and even small errors can propagate through the analysis. But the payoff - a high-quality reference genome for a fascinating species - is well worth the effort. In many ways, it feels similar to how early cartographers must have felt while mapping unfamiliar lands, slowly piecing together the unknown to create a reliable guide for future explorers.
Why this genome matters ?
This reference genome is more than a dataset; it is a powerful resource for discovery. Unlike previous studies that focused primarily on non-migratory resident species at a single elevation, the Black Redstart genome allows us to explore a species that performs short-term altitudinal migration. This opens the door to investigate the molecular basis of physiological acclimation associated with seasonal elevation changes, providing unique insights into how birds cope with rapidly shifting environmental conditions. Ultimately, this reference genome will hopefully transform the Black Redstart into a model system for understanding both high-altitude adaptation and the genetic foundations of flexible migratory strategies in birds.
Looking forward
This project is just the first step. We hope other researchers will use this genome as a resource to explore the Black Redstart’s biology in novel ways. Meanwhile, our team is already investigating gene expression changes across populations of Black Redstart at different altitudes to uncover the molecular strategies these birds use to survive extreme environments.
Building the first reference genome for the Black Redstart has been a journey full of discovery, problem-solving, and excitement. It combined fieldwork on a high-altitude plateau, careful maintenance of sample integrity, state-of-the-art DNA sequencing, and computational genomics, all driven by curiosity about a small bird with a remarkable life story. We are thrilled to share this genome with the scientific community and can’t wait to see the insights it will inspire about evolution, and species adaptation. This genome is just the beginning, these birds and the mountains still hold many secrets, and we hope this map (reference genome) will make navigation easier and exciting.
Follow the Topic
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
Publishing Model: Open Access
Deadline: Apr 17, 2026
Invertebrate omics
Publishing Model: Open Access
Deadline: May 08, 2026
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in