Insights into mechanisms of malaria infection may enable new vaccine approaches
Published in Microbiology, Biomedical Research, and Immunology

While Plasmodium falciparum accounts for most malaria morbidity and mortality worldwide, around 3 billion people are also at risk of P. vivax malaria globally and there is a growing recognition that it is also a major cause of severe malaria and chronic illness. Despite initial gains in reducing malaria in the early 2000s, progress has stalled since 2015. Furthermore, the impacts of climate change are expected to substantially exacerbate malaria in coming years.
New vaccines commencing implementation in young children in Africa (RTS,S and R21) have moderate efficacy, but require annual boosters due to waning immunity and are not active against P. vivax. Therefore, novel vaccines are urgently needed, informed by new knowledge of mechanisms of malaria infection and immunity.
Apical membrane protein 1: Key role in infection and attractive vaccine candidate.
P. falciparum and P. vivax are thought to have independently evolved from primate to human hosts and have adopted alternative erythrocyte invasion mechanisms, ligands and receptors. Apical membrane antigen 1 (AMA1) plays a key role in the infection (or invasion) of human erythrocytes and is one of few proteins shared between P. vivax and P. falciparum. Although the sequence varies between P. falciparum and P. vivax (approx 50% sequence identity), it is thought to play a similar important role in the infection erythrocytes and the development of malaria disease.
We studied AMA1 as an attractive target for vaccines that could be active against P. falciparum and P. vivax. We conducted a series of studies to better understand the specific functions of AMA1 to help inform strategies to develop highly protective vaccines that target AMA1 in both types of malaria.
Previously it was established that AMA1 interacts with a specific loop of the protein Rhoptry Neck-2 (RON2), and it was thought that this interaction was essential for invasion. Our new findings provide evidence that PfAMA1 and PvAMA1 use two or more binding interactions for infection of erythrocytes (Figure 1). Mutation of specific amino acids in PfAMA1 that are known to interact with RON2 did not impact on erythrocyte infection by P. falciparum. Similarly, mutation of key RON2-binding residues in PvAMA1 did not inhibit infection. Our findings indicate that the AMA1–RON2-loop interaction is not essential for infection and additional AMA1 interactions are involved.
Developing vaccines based on AMA1
PfAMA1, produced as a full-length recombinant protein, has been previously evaluated as a vaccine candidate, with limited efficacy. Designing vaccines that target key functional epitopes of AMA1 may be crucial to achieving substantial protective efficacy. Our findings suggest that vaccines and therapeutics based on AMA1 will need to be broader than targeting only the AMA1–RON2 interaction. Targeting multiple AMA1 interactions involved in invasion may enable vaccines that generate more potent inhibitory antibodies and address the capacity of AMA1 for immune evasion through sequence polymorphisms. Our studies revealed one potential approach - we found that the Loop1a peptide of PvAMA1 blocked the binding of PvAMA1 to erythrocytes suggesting it may be involved in erythrocyte binding to enable infection. PvAMA1 Loop1a has no polymorphisms in contrast to other PvAMA1 loops and may be an attractive vaccine target.
Our findings of specific residues for invasion function and species divergence and conservation can inform novel vaccines and therapeutics against malaria caused by both species, including the potential for cross-species vaccines.
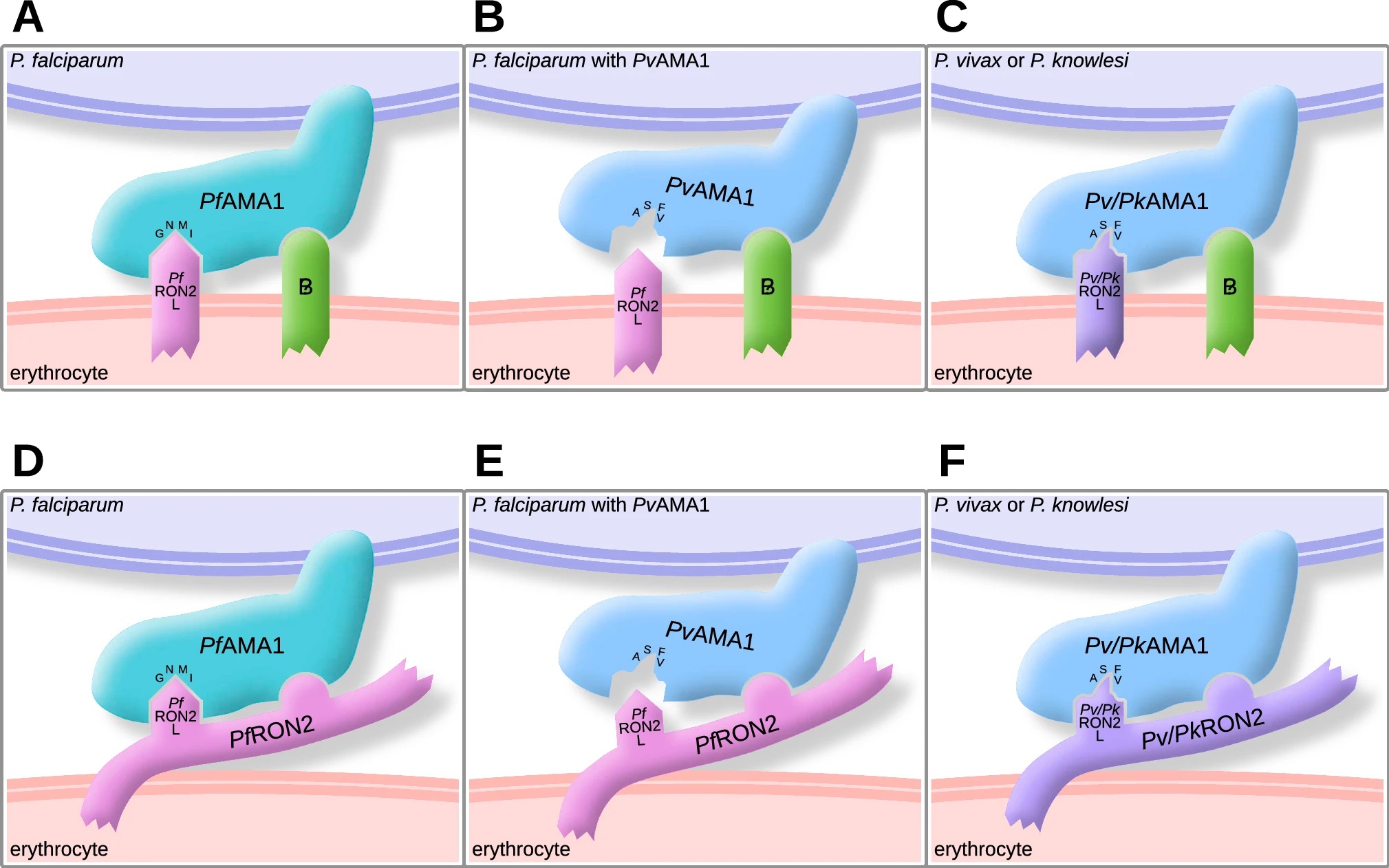
Figure 1. Models of possible AMA1 binding interactions for invasion. AMA1 is present on the merozoite surface and the RON2-loop (labelled as RON2L) is present on the erythrocyte surface. AMA1 is known to bind to the RON2-loop and new data suggest that there is an additional binding interaction of AMA1. In one model (panels A–C), AMA1 binds to Receptor B, which is a proposed molecule on the erythrocyte surface, yet to be identified, that acts a second receptor for AMA1 binding. In the alternative model (panels D–E), the second binding interaction of AMA1 is to a second site on RON2 that is separate to the RON2-loop. A, D. In P. falciparum parasites, PfAMA1 binds to the PfRON2-loop (PfRON2L) and either Receptor B (A) or another region of PfRON2 (D) to mediate invasion. Key amino acids of PfAMA1 Loop1E for binding to RON2 are indicated (G, N, M, and I), as identified in this study. B, E. In genetically modified P. falciparum parasites expressing PvAMA1, PvAMA1 does not bind to the PfRON2-loop, but binds to either Receptor B (B) or another region of PfRON2 (E), allowing invasion to occur. The lack of binding of PvAMA1 to the PfRON2-loop is due to differences in key amino acids of AMA1 Loop1E between P. vivax and P. falciparum (ASFV for PvAMA1 and GNMI for PfAMA1). C, F. In P. vivax and P. knowlesi parasites, PvAMA1 and PkAMA1 can bind to the PvRON2-loop or PkRON2-loop, as well as Receptor B (C) or another region on PvRON2 or PkRON2 (F). There is conservation of AMA1-RON2-loop binding between the two species. The amino acid sequences of the β-hairpin loop of RON2 are identical between P. vivax and P. knowlesi. Of the key RON2-interacting amino acids of AMA1 Loop1E, only one amino acid differs between P. vivax and P. knowlesi. The native conformation and surface orientation of RON2 is not currently fully defined. From Drew DR, et al, CMLS 2023 https://link.springer.com/article/10.1007/s00018-023-04712-z
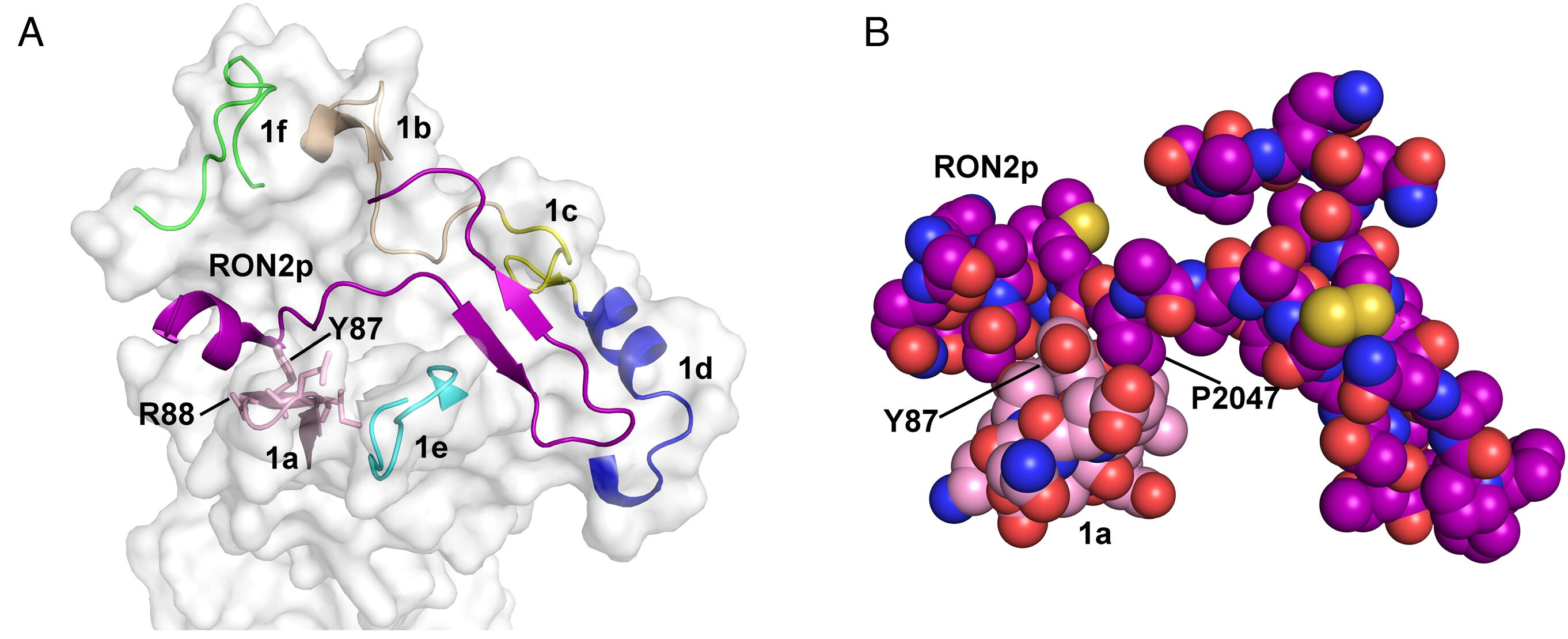
Figure 2: (A) Molecular surface of PvAMA1 with PvRON2L-interacting loops (labeled 1a–1f) (23) is shown in ribbon format. PvAMA1 Loops 1a (pink), 1b (tan), 1c (yellow), 1d (blue), 1e (cyan) and 1f (green) surround the bound RON2L peptide (purple). Side chains of PvAMA1 Loop 1a that contact the PvRON2L ligand are shown in stick representation; Arg 88 and Tyr 87 are labeled. (B) Space-filling representation of PvAMA1 Loop 1a (carbons colored pink) and the PvRON2L peptide (carbons colored purple) showing the intimate association of the loop with the peptide. Residues Tyr 87 from PvAMA1 and Pro 2047 from RON2L are labeled. Oxygen atoms are colored red, nitrogen atoms blue and sulfur atoms yellow. From Lee SK et al, PNAS 2023. https://doi.org/10.1073/pnas.2215003120
Publication references:
Drew DR, Wilson DW, Weiss GE, Yeoh LM, G Henshall I, Crabb BS, Dutta S, Gilson PR, Beeson JG. Defining species-specific and conserved interactions of apical membrane protein 1 during erythrocyte invasion in malaria to inform multi-species vaccines. Cell Mol Life Sci. 2023 Feb 27;80(3):74. https://link.springer.com/article/10.1007/s00018-023-04712-z
Lee SK, Low LM, Andersen JF, Yeoh LM, Valenzuela Leon PC, Drew DR, Doehl JSP, Calvo E, Miller LH, Beeson JG, Gunalan K. The direct binding of Plasmodium vivax AMA1 to erythrocytes defines a RON2-independent invasion pathway. Proc Natl Acad Sci U S A. 2023 Jan 3;120(1):e2215003120. https://doi.org/10.1073/pnas.2215003120
Follow the Topic
-
Cellular and Molecular Life Sciences

Cellular and Molecular Life Sciences (CMLS) is a multidisciplinary Open Access journal covering the latest aspects of biological and biomedical research.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in