Introducing scSemiProfiler: A Deep Generative AI and Active Learning Framework Providing Cost-effective Single-cell Data
Published in Bioengineering & Biotechnology, Protocols & Methods, and Computational Sciences

Single-cell sequencing is a breakthrough in biological research, offering deep insights into cellular complexity by providing cell state information at the individual cell level. This technology is essential for identifying and characterizing various cellular subpopulations in patient samples, driving significant advances in biomarker discovery and personalized therapies. However, its prohibitive cost substantially limits its application in large-scale studies, such as complex disease cohorts. In this work, we leverage deep generative AI and active learning to reduce costs substantially, enabling cost-effective semi-profiling for large-scale single-cell studies.
The cost challenge
Single-cell sequencing is revolutionary but also prohibitively expensive. For instance, according to 2023 estimates from the McGill University Health Centre, sequencing 20,000 cells can cost approximately $6,000, making it impractical for large-scale research projects. To mitigate costs, researchers have used deconvolution methods, such as CIBERSORTx, to infer cell type proportions from more affordable bulk sequencing data. While useful, these methods can provide only cell-population level estimations and lack the detailed resolution needed for single-cell level analyses, which is crucial for understanding complex diseases and also therapeutic explorations.
Developing a “semi-profiling” approach
To leverage the information from affordable bulk data and provide more cost-effective single-cell data, we designed a deep generative AI method to "semi-profile" disease cohorts at the single-cell level accurately and efficiently, in combination with active learning. The entire framework can be broadly categorized into two major parts, both critical for delivering accurate and effective semi-profiling: selecting the best representative single-cell samples (representative selection) and in silico inferring the target single-cell data from the bulk (in silico inference).
In silico single-cell inference using a deep generative learning model
The most crucial part is that given single-cell reference data, how to deconvolute a bulk sample into a single-cell sample? We approach this question by utilizing the connection between bulk data and single-cell data, which is that bulk data is the average of single-cell data. Therefore, if a model is generating the single-cell data for a target sample in a cohort, that only has bulk data, then the average value of the generated single-cell data should be similar to its bulk data. Based on this intuition, we designed our method: a deep generative learning model first reconstructs the single-cell reference data, and then introduces the target sample’s information by requiring the generated single-cell data to have an average value similar to the target sample’s bulk data.
Representative sample selection using active learning
With the in silico inference method, how to select the representative samples such that their single-cell reference data can maximize the inference performance? An intuitive approach is that we can cluster the samples based on their bulk data and pick the samples that are closest to the cluster centroids. However, often it is hard to decide the number of representative samples in advance. Therefore, we adopted using active learning, a category of algorithm for iteratively selecting the most informative samples to maximize machine learning models’ performance. We select the initial batch of representatives based on clustering of the bulk data, and then for the subsequent rounds of representative selection, an active learning algorithm is used for iterative selection, taking into account the bulk data information, the real-profiled and inferred single-cell data information, and the training difficulties of the models. The newly selected representative samples will be subject to single-cell sequencing and used as new single-cell reference data for the new round of in silico inference step. The advantage of this iterative representative selection approach based on active learning is that the user can check the stop criteria, which evaluates the quality of the semi-profiling, and stop the loop at any time to minimize the cost.
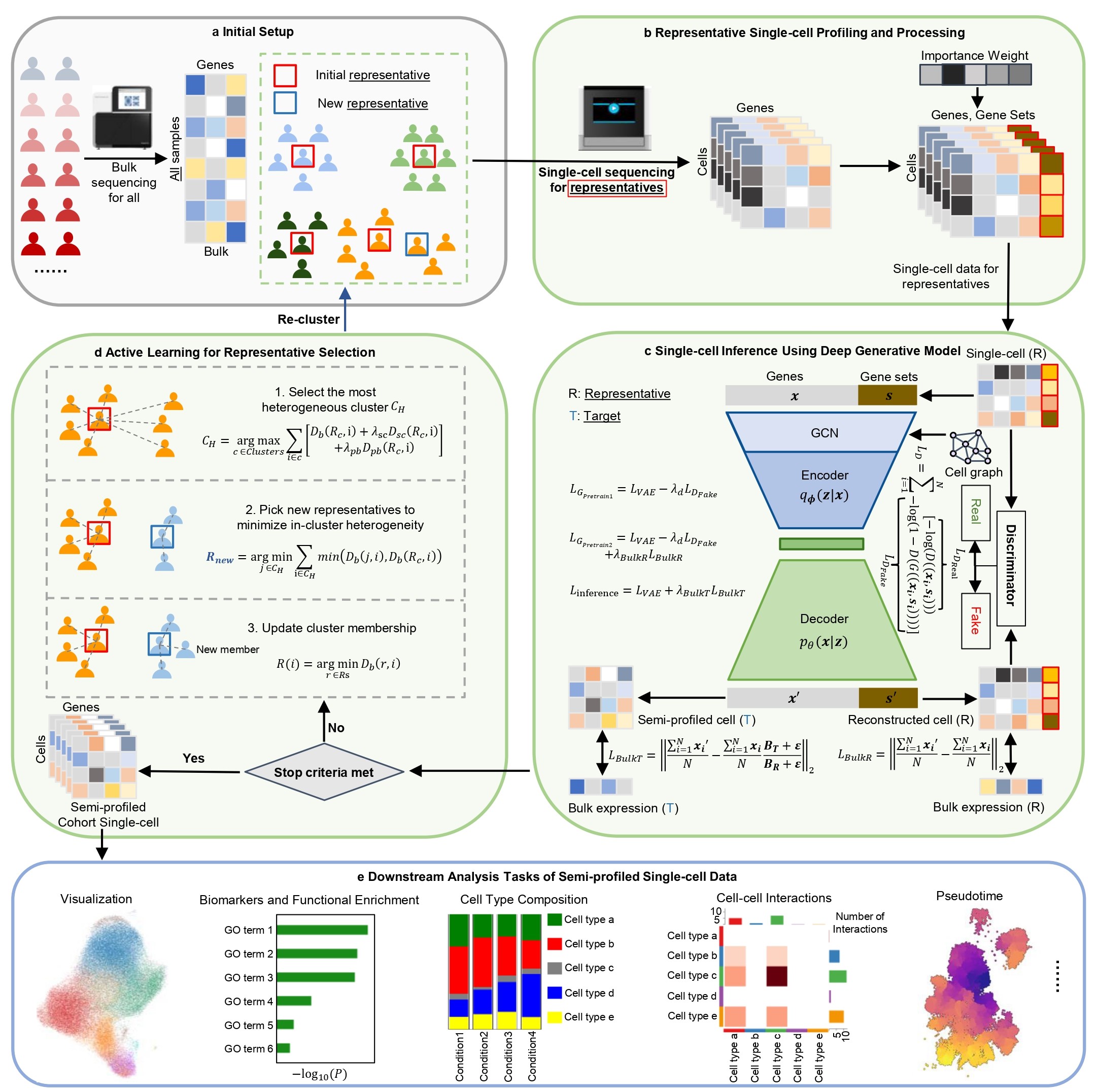
Fig. 1: Overview of the scSemiProfiler method. a Initial setup: Bulk sequencing is performed for the entire cohort. Initial representative samples are selected based on clustering analysis of the bulk data. b Representative sample single-cell sequencing and processing: Single-cell sequencing is conducted for the representative samples. The data is procesed to enhance the performance of deep learning models. c Single-cell inference using deep generative model: Deep generative learning models infer single-cell data for target samples without single-cell data, using bulk data and single-cell data from the representative samples. d Active learning: An active learning algorithm is applied for additional representative sample selection. e Downstream analysis: The semi-profiled dataset can be used for single-cell level analysis, yielding results similar to those obtained with real-profiled datasets.
Validation of scSemiProfiler
We validated scSemiProfiler across diverse datasets, including large disease cohorts with single-cell data for all samples, and cohorts with both single-cell and bulk data for each sample. Results show that scSemiProfiler consistently produces semi-profiled single-cell data closely matching actual single-cell datasets in all datasets. This accuracy is critical for reliable downstream analyses, including visualizations, biomarker discovery, deconvolution, enrichment analysis, cell-cell interaction analysis, pseudotime, etc., making scSemiProfiler a robust tool for large-scale studies at an affordable cost.
Conclusion
scSemiProfiler is a significant advancement in single-cell sequencing, offering a scalable and cost-effective solution for detailed cellular analysis. By overcoming the cost barrier, scSemiProfiler enables researchers to conduct large-scale, high-resolution studies previously unattainable. We invite the research community to explore scSemiProfiler and consider its application in their studies. For more information, read our publication in Nature Communications.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in