Explore the Research
KDM6B protects T-ALL cells from NOTCH1-induced oncogenic stress - Leukemia
Leukemia - KDM6B protects T-ALL cells from NOTCH1-induced oncogenic stress
Where did we start?
T-cell acute lymphoblastic leukemia (T-ALL) is one of the most aggressive blood cancers especially in the adult population (1). With recent sequencing advancements in the genomics era, the driver and cooperating mutations of adult T-ALL have largely been identified (2), revealing a high proportion of genetic mutations that affect epigenetic regulation. However, therapies targeting the pathogenic epigenome are still quite limited in T-ALL.
In a previous study by our group, we demonstrated that the absence of Kdm6b, an epigenetic regulator which serves as a H3K27me3 histone demethylase enzyme, in leukemia cells renders them at a survival disadvantage in certain cellular contexts (3). When we examined normal hematopoiesis, we found that Kdm6b leads to a competitive disadvantage of mouse hematopoietic stem and progenitor cells, a defect that was only evident under stressful conditions, such as serial bone marrow transplantation or inflammation.
Therefore, these results motivated us to explore the role of Kdm6b in T-ALL disease initiation and maintenance and to translate those findings in primary T-ALL patient samples.
What did we do?
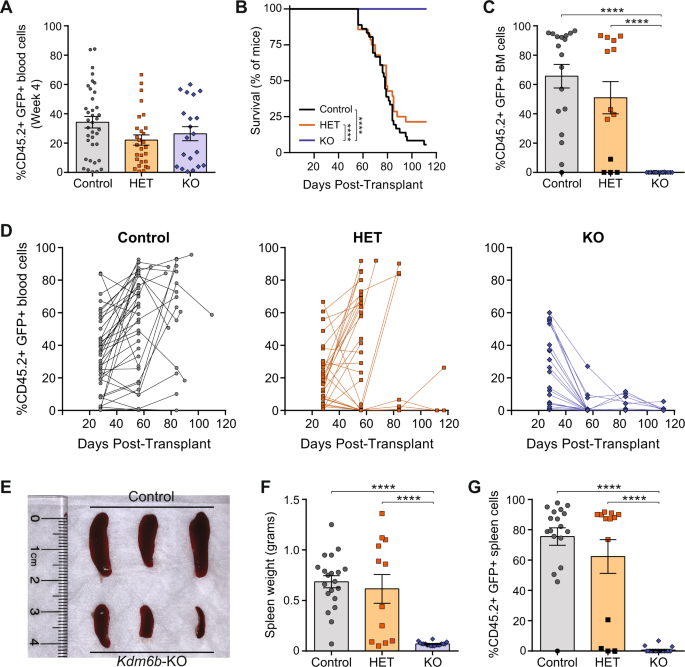
To examine the role of Kdm6b in T-ALL development, we used a retroviral model of NOTCH1 intracellular domain (NICD) expression that recapitulates many of the human pathologies of NOTCH1-mutant T-ALL (4). Using our genetic mouse models, Sca-1 enriched bone marrow cells from Kdm6b-WT, Kdm6b-HET, and Kdm6b-KO adult mice was transduced with a retrovirus expressing Notch1 Intracellular Domain (NICD). Transduced cells were transplanted into irradiated mice. Serial analysis of the peripheral blood and BM of transplanted mice was performed to examine T-ALL disease progression and disease burden at the time of sacrifice.
To investigate the translational potential of our findings, KDM6B was inactivated in primary human patient T-ALL cells by CRISPR/Cas9 gene targeting (5). A gRNA that targets the inert AAVS1 locus was used as a negative control. gRNA/Cas9 ribonucleoprotein (RNP) complexes were nucleofected into primary human T-ALL cells and 48-hours post-nucleofection, these nucleofected cells were xenografted into NSG mice. Analysis of the variant allele fraction (VAF) of CRISPR targeted cells was performed over time to reveal whether targeted T-ALL cells displays a survival difference compared to the control group.
What results have we obtained?
While there was robust engraftment in all groups at four weeks post-transplant, mice receiving Kdm6b-KO NICD-GFP+ cells did not propagate T-ALL within the observation period with no evidence of residual T-ALL in the BM. Serial analysis of the peripheral blood showed T-ALL cells were not sustained in the absence of Kdm6b. Analysis of moribund control and Kdm6b-HET mice displayed significant T-ALL cell burden in all hematopoietic organs. Moreover, mice that received the Kdm6b-KO cells showed no evidence of T-ALL in the BM and spleen at the time of sacrifice. These data suggested NOTCH1-mutant T-ALL cells require some level of Kdm6b for their propagation.
Moving onto our PDX model, analysis of the variant allele fraction (VAF) of CRISPR targeted cells over the experimental time course revealed a striking trend and segregated the response of patient cells into two distinct entities, “responders” and “non-responders”. The VAF of control AAVS1 edited alleles remained relatively consistent. However, a similar comparison of the VAF of KDM6B edited alleles showed that while the targeted cells were efficiently retained in the non-responder patient specimens, the targeted cells were rapidly outcompeted over time in the responder T-ALL patient cells.
We attempted to integrate transcriptome and mutation profile to identify why only a subset of T-ALL patient specimens were sensitive to KDM6B inhibition. Mutational patterns observed in KDM6B responders (multiple NOTCH1 mutations, NOTCH1 plus FBXW7 mutations) serve to increase the strength of NOTCH1 signaling in T-ALL cells. We found the “responder” patients had the highest level of NICD expression, conveying strong NOTCH1 signaling.
What is our main conclusion?
This study identifies KDM6B as an essential gene for NOTCH1-mutant T-ALL cells. Mouse hematopoietic progenitor cells deficient for Kdm6b were unable to initiate T-ALL driven by strong NOTCH1 activating mutations, and primary T-ALL cells from human patients targeted for KDM6B loss-of-function by CRISPR/Cas9 gene editing were rapidly outcompeted by non-edited cells. These outcomes suggest KDM6B mutations are never observed in T-ALL patients as this would result in loss of these clones due to competitive disadvantage.
We hypothesize that KDM6B is required in this subgroup of patients to protect the leukemia cells from NOTCH1 oncogene stress-induced apoptosis, although the precise mechanism of this protection conveyed by KDM6B remains to be elucidated.
This study identifies a novel role for KDM6B in maintenance of T-ALL driven by strong NOTCH1 signaling, which opens the possibility for a new class of therapeutic discovery for this subset of T-ALL patients.
References
- Pui CH, Robison LL, and Look AT. Acute lymphoblastic leukaemia. Lancet. 711 2008;371(9617):1030-43.
- Neumann M, Vosberg S, Schlee C, Heesch S, Schwartz S, Gokbuget N, et al. 732 Mutational spectrum of adult T-ALL. Oncotarget. 2015;6(5):2754-66.
- Mallaney C, Ostrander EL, Celik H, Kramer AC, Martens A, Kothari A, et al. Kdm6b 752 regulates context-dependent hematopoietic stem cell self-renewal and leukemogenesis. 753 Leukemia. 2019.
- Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, et al. Exclusive 771 development of T cell neoplasms in mice transplanted with bone marrow expressing 772 activated Notch alleles. J Exp Med. 1996;183(5):2283-91.
- Gundry MC, Brunetti L, Lin A, Mayle AE, Kitano A, Wagner D, et al. Highly Efficient 791 Genome Editing of Murine and Human Hematopoietic Progenitor Cells by 792 CRISPR/Cas9. Cell Rep. 2016;17(5):1453-61.
Follow the Topic
-
Leukemia

This journal publishes high quality, peer reviewed research that covers all aspects of the research and treatment of leukemia and allied diseases. Topics of interest include oncogenes, growth factors, stem cells, leukemia genomics, cell cycle, signal transduction and molecular targets for therapy.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in