Largest SARS-CoV-2 genomic surveillance study in Pakistan reveals critical variant dynamics and regional evolution patterns
Published in Microbiology, Protocols & Methods, and Genetics & Genomics
Background
Pakistan, with over 240 million people, experienced devastating COVID-19 waves but lacked comprehensive genomic surveillance data. Previous studies were limited in scope, covering fewer locations and shorter time periods. Understanding SARS-CoV-2 evolution in South Asia was crucial for global pandemic response, yet this region remained underrepresented in genomic databases.
Key Results
Our analysis revealed several surprising patterns:
A clear handover between variants: Alpha dominated from March through May 2021 (69% of cases in March), while Delta took over from June onwards (91% by September). This wasn't gradual mixing - it was a rapid replacement that mirrored global trends but with Pakistan-specific timing.
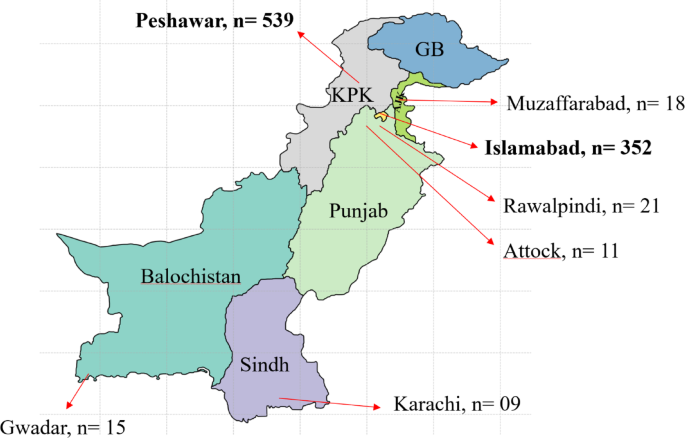
Geographic hotspots told different stories: Peshawar became an Alpha stronghold (62% of samples), likely due to cross-border travel, while Islamabad emerged as Delta's entry point (89% Delta by our sampling period). This suggests different introduction pathways for different variants.
Pakistan's hidden role in global evolution: We discovered that Pakistan harbored 79 cases of the AY.108 lineage - representing about one-third of all known cases worldwide at the time. This was completely unexpected and suggests regional viral evolution we hadn't previously recognized.
Urban transmission networks: Our phylogenetic analysis showed clear transmission chains within cities, with some variants staying localized while others spread rapidly between urban centers.
Why It Matters
This work demonstrates that comprehensive genomic surveillance is possible even with limited resources. Using Oxford Nanopore's portable sequencing technology, we processed samples from distributed collection sites across Pakistan - proving that countries don't need massive infrastructure investments to participate in global genomic surveillance.
More importantly, our findings highlight how much we were missing about global viral evolution. If Pakistan - a country with limited prior sequencing - harbored such a significant proportion of a global lineage, how many other evolutionary patterns are we missing in undersampled regions?
The study also provides a roadmap for pandemic preparedness. Our data showed variant transitions happening 4-6 weeks before major case surges, potentially giving health systems crucial warning time for future outbreaks.
The research establishes Pakistan's position in global SARS-CoV-2 transmission networks and provides a framework for future pandemic response in the region.
Published in: Scientific Reports DOI: https://doi.org/10.1038/s41598-025-12774-1
Follow the Topic
-
Scientific Reports

An open access journal publishing original research from across all areas of the natural sciences, psychology, medicine and engineering.
Related Collections
With Collections, you can get published faster and increase your visibility.
Reproductive Health
Publishing Model: Hybrid
Deadline: Mar 30, 2026
Women’s Health
Publishing Model: Open Access
Deadline: Feb 28, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in