Lesion Segregation
Published in Cancer
The Liver Cancer Evolution (LCE) Consortium has just published our “lesion segregation” manuscript in Nature 1. This has been an exciting and unexpected journey benefiting from elements of curiosity, serendipity and flexibility of funding, built on the solid foundation of a well-designed study with interdisciplinary and international collaboration.
Over the past decade, the sequencing of cancer genomes has provided a goldmine of insights into the mutations that drive the development of cancer, and the processes that generate and shape the patterns of mutation. Duncan Odom recognised that a well-powered, well-controlled model organism system would elegantly complement the many ongoing human cancer studies. At a characteristically grand scale, he set out to re-run liver cancer evolution hundreds of times, controlling genetic background, environment, age, gender and mutagen exposure. This huge project spanned several years: Sarah Aitken, Frances Connor and others from Duncan’s group (CRUK Cambridge Institute) performed the experiments and generated all the primary data, with most sequencing provided by Edinburgh Genomics. For the analysis, Duncan corralled together the Consortium, comprising the labs of Paul Flicek (EMBL-EBI, Cambridge), Núria López-Bigas (IRB, Barcelona), Colin Semple (MRC-HGU, Edinburgh) and my lab (MRC-HGU, Edinburgh).
We ploughed into the rich data with enthusiasm, sharing insights, questions and coordinating analyses with face-to-face meetings in Cambridge, Barcelona, Heidelberg and Edinburgh - and regular Zoom calls (well before the pandemic popularity of Zoom). There are several exciting stories to report from this collaboration but the first finding - lesion segregation - was a completely unexpected discovery.
I heard about “transcriptional mutagenesis” from Paul Doetsch while visiting North Carolina. This is where a damaged DNA nucleotide leads to the production of an activated oncogene protein before the damage has been repaired or resolved into a mutated DNA base pair2,3. Our liver cancer model system, with a single large dose of mutagen, seemed to be a good system in which to look for evidence of transcriptional mutagenesis. The pattern I was looking for was a subset of driver mutations with unusually high variant allele frequency and evidence that the responsible damaged nucleotide was specifically on that gene’s template strand. While this pattern did not seem to be prominent in the data, looking for it raised the questions of when does the lesion become a mutation? And what would happen if the lesion remained unrepaired after replication? Sketching it out suggested that near-by lesions, if not repaired before or during replication, would create clusters of strand-asymmetric mutations. Extrapolating this seemed implausible because it suggested that these clusters of asymmetry should extend to the ends of the chromosomes. Nevertheless, a few lines of R code later I was looking at an xy-scatter plot showing whole X-chromosomes with almost all mutations from T on the forward strand and, in other tumours, X-chromosomes with only mutations from A on the forward strand.
From there it was quickly down the rabbit-hole as we realised we had DNA lesions segregating into separate sister chromatids, resulting in the genome-wide, chromosome-scale phasing of mutations. We could perform analyses that were previously impossible: resolving recombination between sister chromatids, quantifying oncogenic selection, finding known and previously unknown reference genome misassemblies, and measuring transcription coupled repair with the knowledge of which strand the DNA lesions had occurred on. We concluded that DNA damage must survive as lesions for at least one cell division, could it survive for more divisions and create new mutations each generation? We sent Craig Anderson on the mission to look for what seemed like the remote possibility of multi-allelic variation; that is, multiple different genetic changes at the same nucleotide position in the same tumour. He quickly came back to report that it was frequent in our data and then diligently reconfirmed that result using a host of orthogonal approaches. The genetic diversity generated by these unrepaired lesions has profound implications for understanding Darwinian selection in cancers.
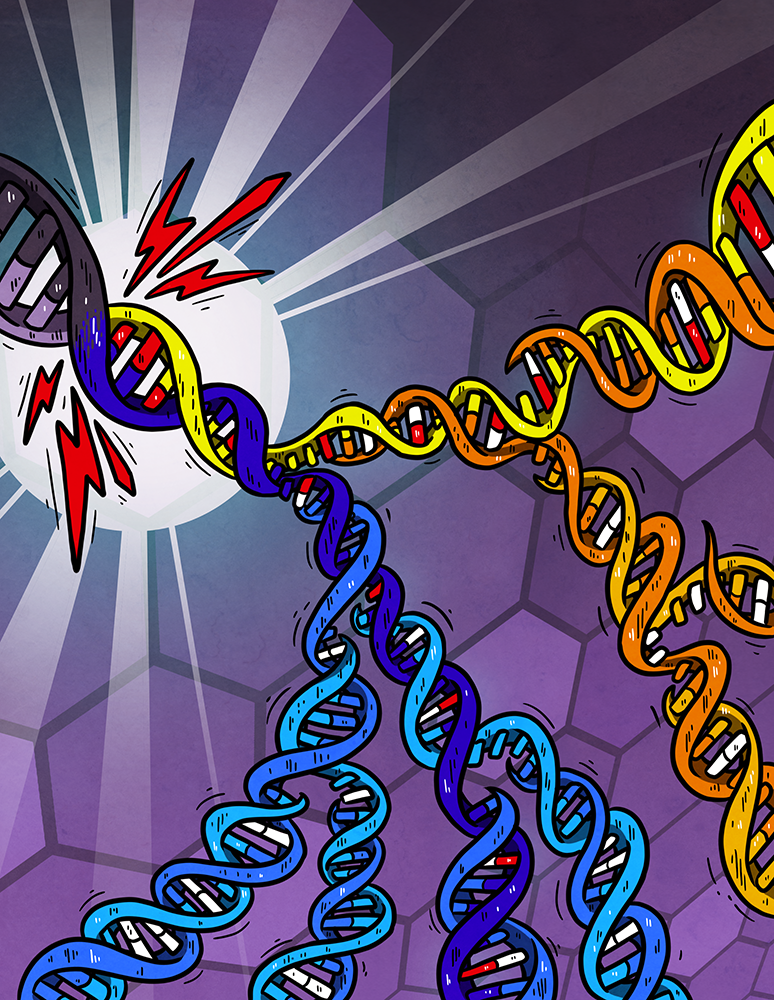
Cartoon illustrating DNA damage (red bases) generated independently on both strands of the DNA helix by a burst of an exogenous mutagen (lightning-bolt). Subsequent replication over the unrepaired damage incorporates the incorrect base (white) opposite damage which is resolved into a mutated DNA base-pair by later replication. Lesions can persist for multiple cell generations generating new combinations of mutation each time they are used as a template for replication. Artwork from Petra Korlevic @petrathepostdoc.
While this whirlwind of analyses was taking place, a brilliant and very timely paper was published by Serena Nik-Zainal’s group, in which they had treated a human derived cell line with a catalogue of known and suspected mutagens 4. Although the mutation loads were much lower than in our carcinogen-induced liver tumours, we could see the characteristic mutational asymmetry patterns of lesion segregation in all the samples that had good statistical power. This demonstrated that lesion segregation occurs in human cells and appears to be a unifying feature of mutagenesis through DNA base damage. The same software we had written to re-analyse Serena’s data was then used to scan the latest public release of the International Cancer Genome Consortium 5 where we could find striking examples of lesion segregation in human cancers. While we think lesion segregation is a general feature of mutagenesis from damaged DNA, the asymmetry patterns are expected to be progressively obscured by recurrent rounds of exposure. This implies that cancers with readily detectable asymmetry were induced by a large single dose of mutagen. An observation that may have epidemiological utility in quantifying exposures.
Chasing down an unexpected observation in a well-controlled experiment led to fundamental insights into the processes of mutagenesis, revealed opportunities for efficient selection in cancer, and has provided new views of mutagenesis and repair processes. Putting the manuscript together was great collaborative fun: me, Sarah and Duncan tag-team writing in GoogleDocs while the wider consortium were feeding in ideas, critiquing and drawing enormous story-boards across the walls of meeting rooms. The core of this work is a synergistic Europe-wide collaboration, but understanding the wider relevance and demonstrating lesion segregation in a clinical setting required the efficient and unencumbered sharing of published research data for which we are very grateful.
- Aitken, S. J. et al. Pervasive lesion segregation shapes cancer genome evolution. Nature (2020) doi:10.1038/s41586-020-2435-1.
- Saxowsky, T. T., Meadows, K. L., Klungland, A. & Doetsch, P. W. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 105, 18877–18882 (2008).
- Brégeon, D. & Doetsch, P. W. Transcriptional mutagenesis: causes and involvement in tumour development. Nat. Rev. Cancer 11, 218–227 (2011).
- Kucab, J. E. et al. A Compendium of Mutational Signatures of Environmental Agents. Cell 0, (2019).
- ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020).
Follow the Topic
-
Nature

A weekly international journal publishing the finest peer-reviewed research in all fields of science and technology on the basis of its originality, importance, interdisciplinary interest, timeliness, accessibility, elegance and surprising conclusions.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in