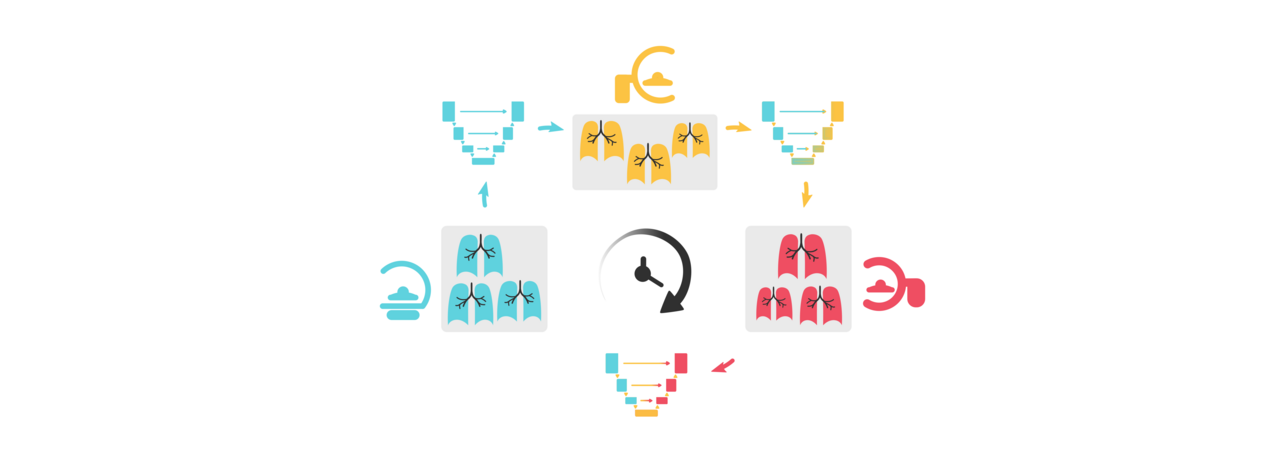
Most research and product development in medical AI focuses on designing static systems for a dynamic clinical world. This disparity often leads to unwanted consequences, such as abrupt drops in performance post-deployment1,2. Over time, as acquisition standards and patient populations change, models trained with an outdated data distribution become unreliable and unsuitable for clinical use.
Continual learning systems are becoming a viable alternative and raising the interest of medical image researchers and regulatory bodies alike3,4. The shift from static to continual learning is a significant development in the design and evaluation of AI solutions which proposes to train models through the entire product lifecycle, even post-deployment. Despite recent advances5-8, research on continual medical image segmentation is still in its infancy, partly due to a lack of a standardized framework that allows for (1) easy benchmarking (2) state-of-the-art performance and (3) a fair comparison between methods9.
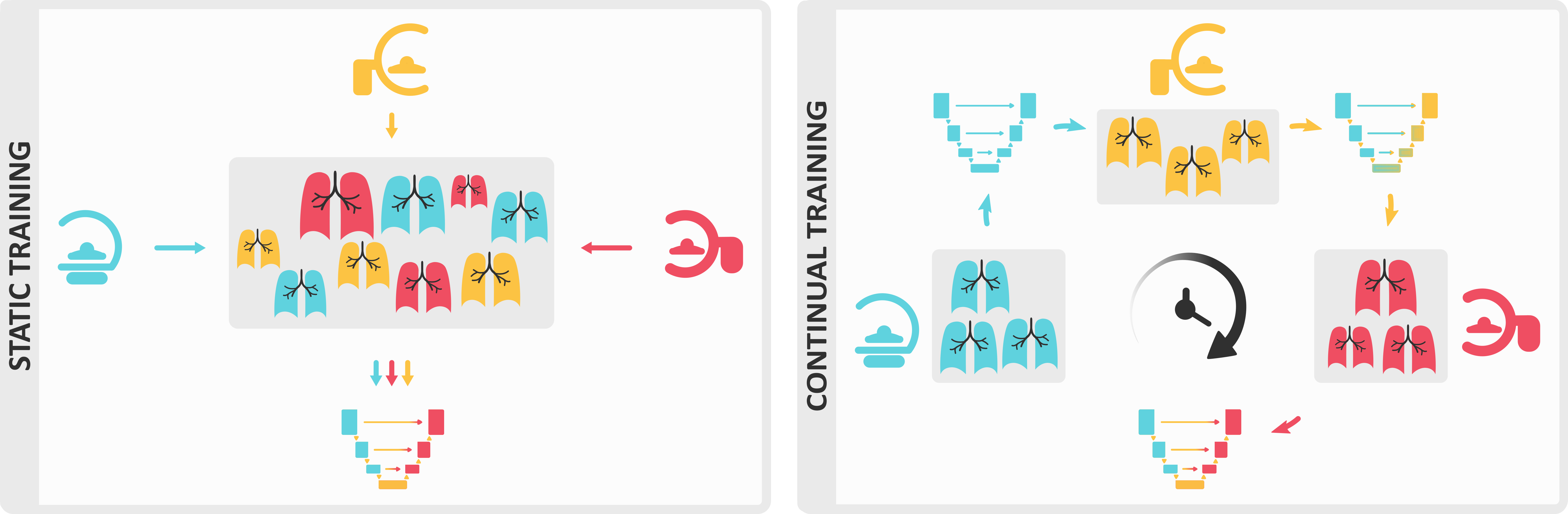
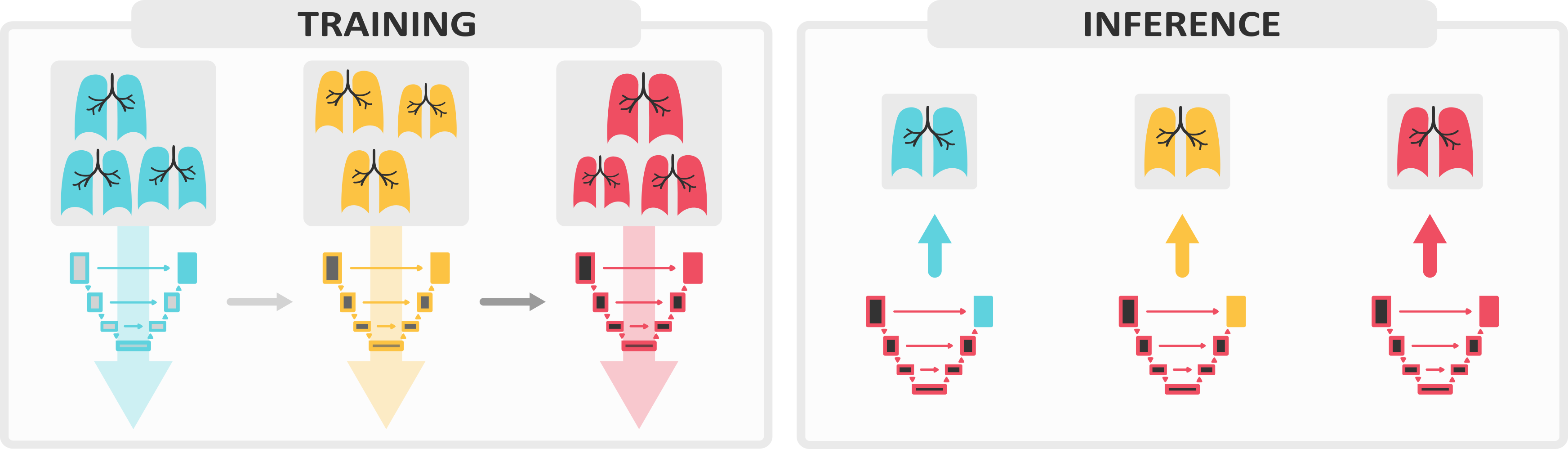
We present Lifelong nnU-Net, a solution for continual medical image segmentation that ensures maximal performance, transferability and ease of use. With only one line of code, users can evaluate the most popular continual learning methods on their data, with guaranteed base segmentation performance. Our library (github.com/MECLabTUDA/Lifelong-nnUNet) integrates with the state-of-the-art nnU-Net10, allowing a wide community of medical image computing enthusiasts to deep into continual learning with minimal set-up.

In our manuscript, we outline the clinical significance of our work and thoroughly describe all technical components designed to ease users into a continual paradigm, such as the practice of maintaining domain-specific heads.
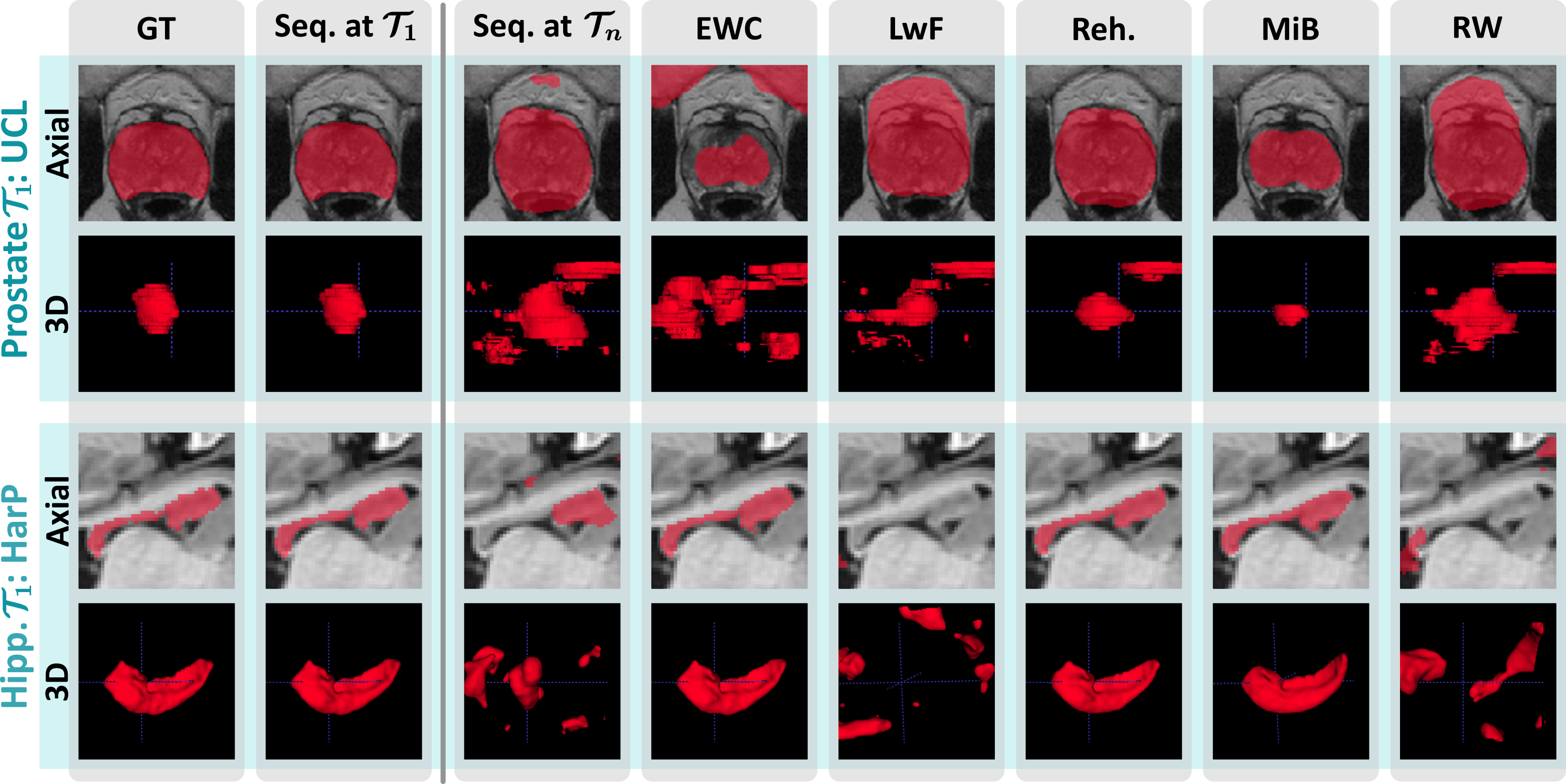
Our evaluation across three use cases chosen for their clinical significance and heterogeneity shines light on the recent progress and open challenges of training segmentation models continuously. It also illustrates relevant aspects of best-practice continual evaluations, such as the importance of task orderings.
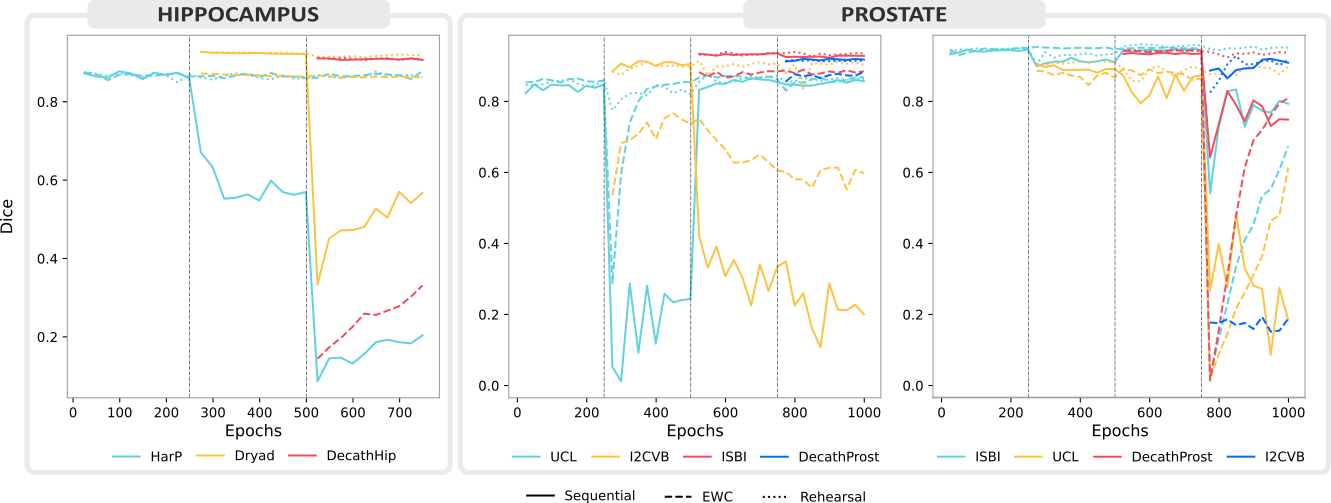
By reporting our results on publicly available data and sharing our code, we hope to drive continual learning research forward in the medical domain.
References
1. Eche, T., Schwartz, L. H., Mokrane, F. Z., & Dercle, L. (2021). Toward Generalizability in the Deployment of Artificial Intelligence in Radiology: Role of Computation Stress Testing to Overcome Underspecification. Radiology: Artificial Intelligence, 3(6), e210097.
2. Liu, X., Glocker, B., McCradden, M. M., Ghassemi, M., Denniston, A. K., & Oakden-Rayner, L. (2022). The medical algorithmic audit. The Lancet Digital Health.
3. Lee, C. S., & Lee, A. Y. (2020). Clinical applications of continual learning machine learning. The Lancet Digital Health, 2(6), e279-e281.
4. Vokinger, K. N., & Gasser, U. (2021). Regulating AI in medicine in the United States and Europe. Nature machine intelligence, 3(9), 738-739.
5. Michieli, U., & Zanuttigh, P. (2019). Incremental learning techniques for semantic segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision Workshops.
6. Cermelli, F., Mancini, M., Bulo, S. R., Ricci, E., & Caputo, B. (2020). Modeling the background for incremental learning in semantic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 9233-9242).
7. Memmel, M., Gonzalez, C., & Mukhopadhyay, A. (2021). Adversarial continual learning for multi-domain hippocampal segmentation. In Domain Adaptation and Representation Transfer, and Affordable Healthcare and AI for Resource Diverse Global Health (pp. 35-45). Springer, Cham.
8. Perkonigg, M., Hofmanninger, J., Herold, C. J., Brink, J. A., Pianykh, O., Prosch, H., & Langs, G. (2021). Dynamic memory to alleviate catastrophic forgetting in continual learning with medical imaging. Nature Communications, 12(1), 1-12.
9. Lomonaco, V., Pellegrini, L., Cossu, A., Carta, A., Graffieti, G., Hayes, T. L., ... & Maltoni, D. (2021). Avalanche: an end-to-end library for continual learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 3600-3610).
10. Isensee, F., Jaeger, P. F., Kohl, S. A., Petersen, J., & Maier-Hein, K. H. (2021). nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods, 18(2), 203-211.
Follow the Topic
-
Scientific Reports

An open access journal publishing original research from across all areas of the natural sciences, psychology, medicine and engineering.
Related Collections
With Collections, you can get published faster and increase your visibility.
Reproductive Health
Publishing Model: Hybrid
Deadline: Mar 30, 2026
Women’s Health
Publishing Model: Open Access
Deadline: Feb 28, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in