
Ubiquitination & phosphorylation
The attachment of a phosphate group to the hydroxyl group of serine, threonine, and tyrosine residues in proteins is well-established. In contrast, ubiquitination is a more complex modification, conjugating to lysine, serine and threonine residues. However, evidence for tyrosine ubiquitination remained elusive despite a long-standing search by the research community. Through a combination of serendipity and methodological advancements, using buffers at a higher pH, tyrosine ubiquitination was identified 46 years after the discovery of tyrosine phosphorylation.
Both ubiquitination and phosphorylation regulate a wide range of cellular processes in eukaryotes and have garnered attention in drug development. The discovery of tyrosine phosphorylation, made 46 years ago, resulted from the repeated use of a homemade buffer with a lower pH. Phosphorylation, in particular, has led to the discovery of more than 40 FDA-approved drugs, such as Gleevec (imatinib), which inhibits the Abl tyrosine kinase. Similarly, targeted protein degradation (TPD) technologies, powered by diverse ubiquitin-related mechanisms, are now advancing through clinical trials, offering promising therapeutic potential for conditions such as cancer.
Identification of tyrosine ubiquitination
Protein ubiquitination is an important and complex cellular regulatory mechanism, playing a key role in signal transduction, immune responses, and cancer development. However, for a long time, ubiquitination has been considered to primarily occur on lysine, with lysine ubiquitination being nearly synonymous with ubiquitination itself. There has been ongoing speculation and a prolonged search for evidence of tyrosine ubiquitination, but clear evidence has been lacking, potentially due to the difficulty of capturing it using traditional methods. The presence of tyrosine ubiquitination in cells has remained an unresolved long-term mystery (Figure 1). Despite recent discoveries and reports of non-lysine ubiquitination, such as threonine and lipopolysaccharide (LPS) ubiquitination, the field has gradually reached a consensus that lysine ubiquitination may represent only a snap of the broader landscape of ubiquitination.
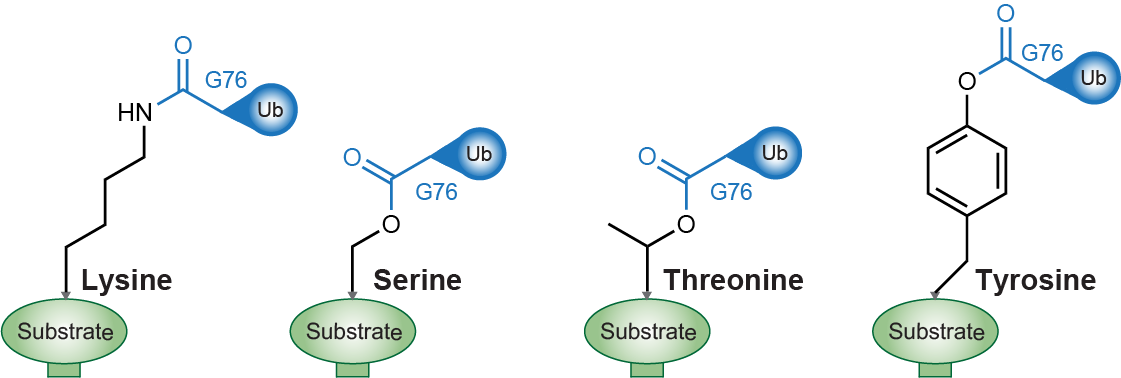
Figure 1. Representative protein ubiquitin conjugations, from left to right: known lysine ubiquitination, serine ubiquitination, threonine ubiquitination and the unknown tyrosine ubiquitination.
The intention of studying protein ubiquitination through the entry point of E2-ubiquitin conjugating enzymes, as the name suggests, plays an important role in determining ubiquitin conjugation. Our goal as chemical biologists is to develop tools that bridge the gap between chemistry and biology. We believe that ubiquitin-based chemical biology tools are of great value for in-depth studies of the ubiquitin landscape. We are intrigued by the complex ubiquitin conjugations in cells, which yield a large smear of polyubiquitin bands, whether through gel-based assays or mass spectrometry. This complexity may be partially resolved by defining specific E2-specific pathways, with a manageable number of E2s (over 30 in humans) and their subsequent conjugations (Figure. 2).
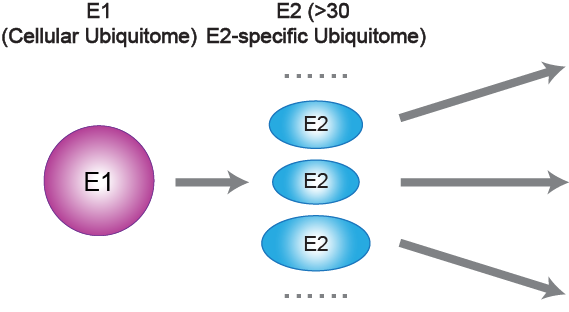
Figure 2. The entire ubiquitin pathway (entire ubiquitome) can be further subdivided into more than 30 E2 sub-pathways (E2-specific ubiquitome) based on E2 enzymes.
We developed an fusion E2-Ub-R74G profiling (FUSEP) strategy that targets the unique chemical properties of non-lysine ubiquitination and optimizes sample preparation, detection, and analysis methods for mass spectrometry. The design of FUSEP probes containing the Ub-R74G mutation allows them to label E2 enzyme substrate proteins in live cells. After trypsin digestion for mass spectrometry, the probe leaves a distinctive LGGG residue (E2-specific ubiquitome) at the modification site, distinguishing it from the GG residues (entire ubiquitome) generated by endogenous ubiquitin modifications (Figure 3). This design aims to enhance ease of use for interested colleagues, with minimal requirements for typical cell culture conditions and molecular biology platforms for transfection. Further requirements include typical gel-based analysis and sample preparation for mass spectrometry analysis.
The discovery of tyrosine ubiquitination was an accidental finding during an unanticipated experiment, where we simply wanted to observe the behavior of UBE2D3 FUSEP probes treated with a higher pH buffer containing hydroxylamine (NH2OH), inspired by a prior illuminating perspective article published in Nature Chemical Biology. Further unexpected results from the mass spectrometry trial led to the observation of an over 8-fold decrease in human Cullin-1 (CUL1), potentially hinting at tyrosine ubiquitination sites on a highly conserved tyrosine residue. This led to and drove further extensive experimental attempts.
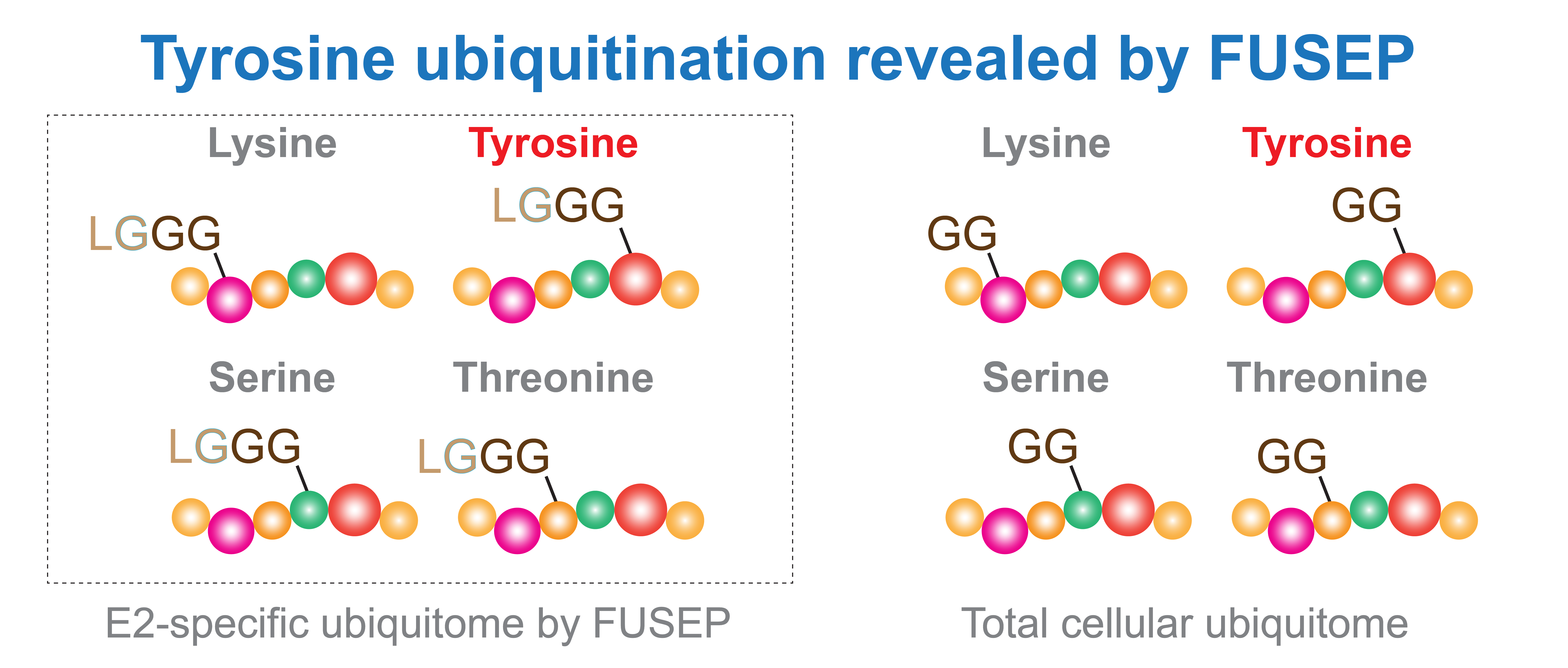
Figure 3. Fusion E2-Ub-R74G profiling (FUSEP) enabled the analysis of the E2-specific ubiquitome, separate from the total cellular ubiquitome, and identified evidence of tyrosine ubiquitination using a higher pH buffer.
Tyrosine ubiquitination and beyond
We used FUSEP technology to reveal the existence of tyrosine ubiquitination at multiple sites on several important proteins, including CUL1, CAND1, HOIL1, HOIP and OGT, in human cells. For example, in the SCF (SKP-CUL1-F-box) E3 complex, FUSEP uncovered tyrosine ubiquitination at multiple positions on the CUL1 protein, particularly at the highly conserved tyrosine residue at position 99. This suggests that, compared to traditional lysine ubiquitination, tyrosine ubiquitination may exhibit unique chemical and biological properties in regulating CUL1 activity. This finding supports FUSEP as an effective tool for enabling in-depth studies of E2-specific lysine and non-lysine ubiquitination conjugations in situ, providing different perspectives to expand our understanding of ubiquitin conjugations.
The existence of tyrosine ubiquitination, which has been largely unknown until now, may raise numerous subsequent questions regarding its biological functions and molecular mechanisms. This could potentially open new avenues for collaborative research efforts. Given the low chemical stability and the still unclear biological function of non-lysine ubiquitination, this area remains in the very early stages. As discussed in the Nature Chemical Biology perspective “A new dawn beyond lysine ubiquitination” and the Nature Structural & Molecular Biology comment “Just how big is the ubiquitin system?”, technological advancements are expected to drive future progress in this field, attracting researchers from diverse technical backgrounds and areas of expertise.
Link to the paper: https://www.nature.com/articles/s41589-024-01809-9
Follow the Topic
-
Nature Chemical Biology

An international monthly journal that provides a high-visibility forum for the chemical biology community, combining the scientific ideas and approaches of chemistry, biology and allied disciplines to understand and manipulate biological systems with molecular precision.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in