From interactions to networks
The RNP-MaP technology evolved from a desire to rapidly investigate novel RNA-protein, or ribonucleoprotein, complexes (RNPs) in a straightforward way. I have been studying RNPs my entire research career, and the prior inability to quickly detect all or most protein binding to a given cellular RNA had really stymied the whole field from pursuing RNPs with unknown function. My dissertation focused on how the RNA-binding protein Pumilio regulates mRNA translation and stability. Near the end of my graduate work, I observed how the protein Nanos could clamp Pumilio onto an RNA and alter the sequences Pumilio could recognize. Nanos expanded the potential list of Pumilio targets from 20% to 70% of the fly transcriptome, and that is the result of a single protein-protein interaction! This Nanos-Pumilio mini-network explained why true targets of proteins like Pumilio are hard to predict based on their individual binding affinities alone, because the target sequence depends on accessory protein-protein interaction networks. The basic idea of RNP-MaP was born from my interest in characterizing these RNP networks.
In search of a sequencing readout
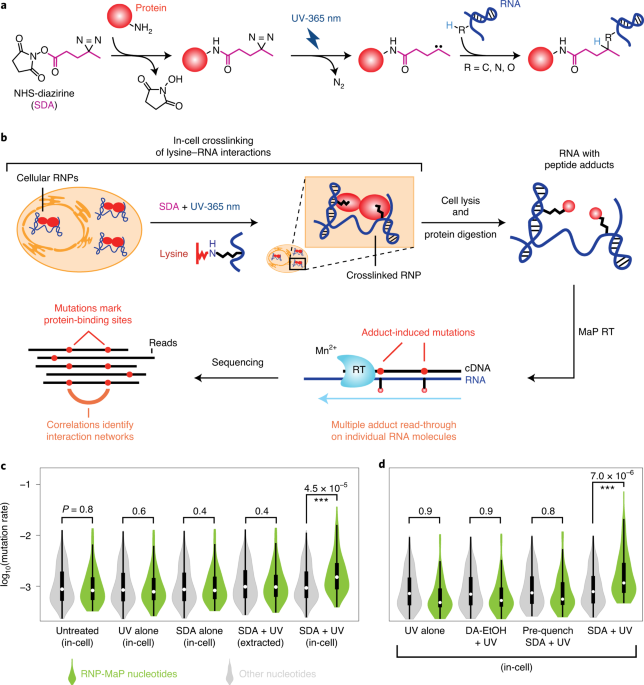
Around that time, every conference featured speakers appending “-seq” onto their RNA discovery technologies, and I knew that whatever training I sought had to include the integration of massively-parallel sequencing and bioinformatics. I decided to join the laboratory of Kevin Weeks, which had recently developed correlated chemical probing and mutational profiling (MaP) to measure through-space RNA-RNA interactions and RNA structure using SHAPE and other chemical probes. I thought that if it were possible to “MaP” intramolecular RNA-RNA interaction networks, why couldn’t I do the same for protein-protein communication on RNA? I pitched the idea in one of my first project meetings with Kevin, originally combining MaP with photo-activatable ribonucleoside labeling and crosslinking. Kevin thought using a chemical probe would make for a simpler approach. I made a short list of heterobifunctional compounds that could theoretically crosslink RNA and protein, and that list included SDA.

Team stare-ology
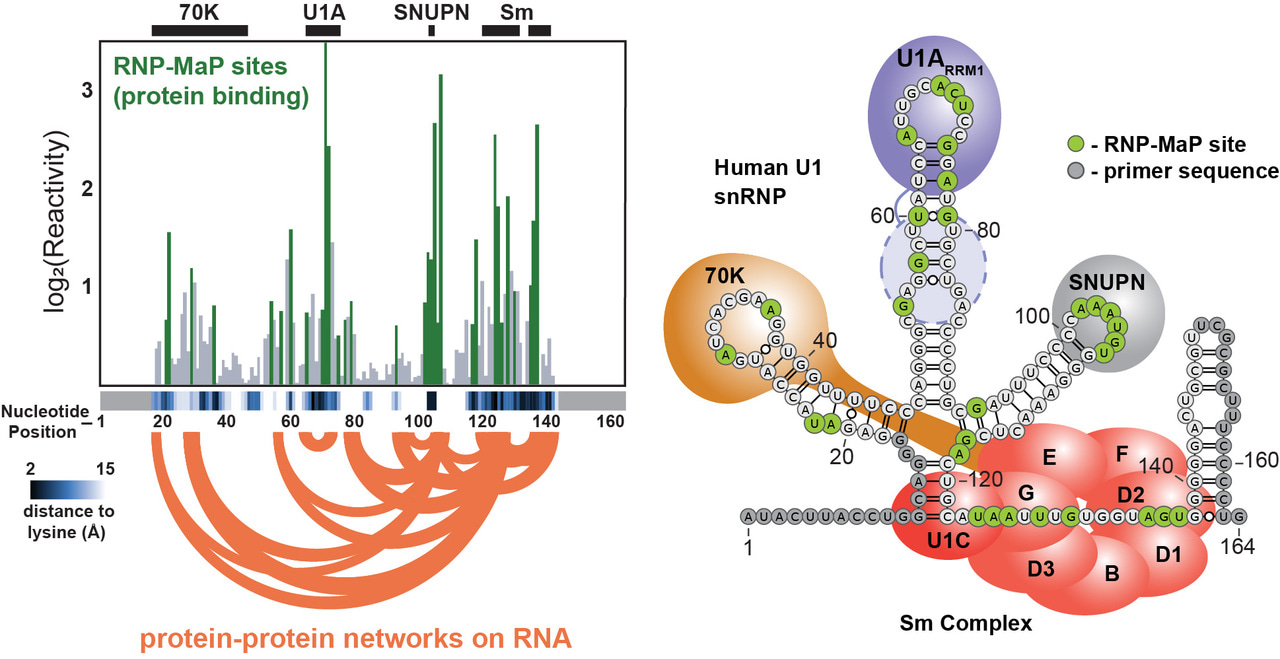
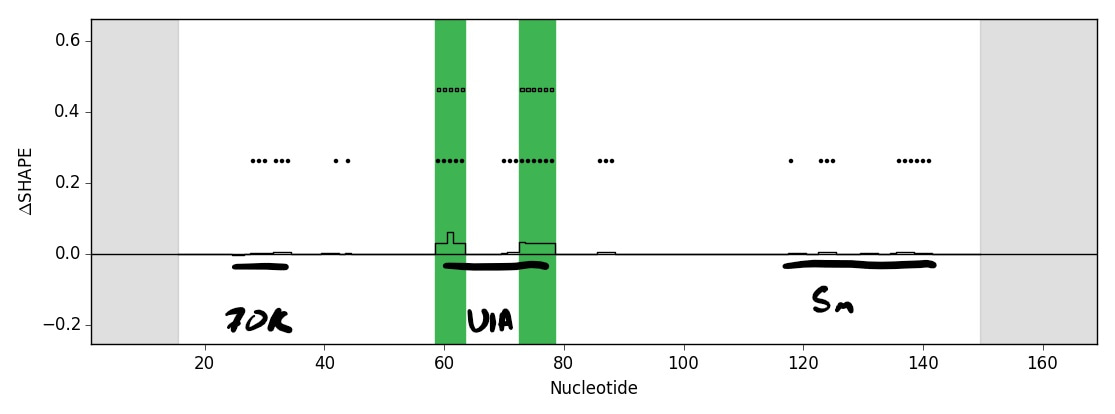
I was able to rapidly test the feasibility of each crosslinker based on existing technologies in the lab. A graduate student, Matt Smola, had recently developed ΔSHAPE to identify protein binding sites in cells. His MaP strategy and analysis pipeline could be adapted for my RNP crosslinkers. SDA was the only compound that had any signal, but it appeared very sparse across the U1 small nuclear RNA that I was using as a model system. I printed out the result of my first SDA trial and showed it to Matt, expressing my concern. Matt prepared to be disappointed and then did a double-take, bringing the paper closer to his face and scrutinizing the data. Matt pointed out that, while small, signal peaks mapped exactly to the known 70K, U1A, and Sm protein binding sites on U1 (my RNP-MaP strategy worked!). The thresholding and scaling in the existing pipeline were specific for SHAPE reactivity and made the SDA signal appear less significant in the original output. By working together to spot the true signal (we call this process “stare-ology” in the lab), I knew RNP-MaP was worth developing further.
A new road-MaP for RNA biology
After that initial result, I validated the approach on multiple RNPs of known structure and created signal thresholds based on empirical measurements of reactivity distributions relative to (RNA) nucleotide-(protein) lysine distances. Another postdoc, Tony Mustoe, updated the correlated chemical probing analysis pipeline to a G-test framework, which further improved RNP-MaP network detection. Ultimately, our RNP-MaP technology could reliably identify the locations of protein binding sites and protein-protein interaction network hubs critical to the functions of multiple RNAs, without any other pre-existing knowledge. I characterized an expansive interaction network on the XIST long noncoding RNA critical for assembly into a nuclear compartment. Based on our analysis of multiple RNAs, it became clear that protein interaction networks can be conserved between RNAs even when their nucleotide recognition sequences differ, revealing a mode of evolutionary adaptation through protein-RNA interactions. RNP-MaP ultimately enables quantitative comparison of RNP interaction networks between cell states at nucleotide resolution: RNP-MaP will be a boon for discovering and understanding diverse functions of coding and noncoding RNAs.
Follow the Topic
-
Nature Biotechnology

A monthly journal covering the science and business of biotechnology, with new concepts in technology/methodology of relevance to the biological, biomedical, agricultural and environmental sciences.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in