Mapping spruce bark degradation over time identifies resin acids as key drivers of microbial diversity
Published in Ecology & Evolution, Microbiology, and Plant Science

Trees can be seen as huge reservoirs of carbohydrates that would be an attractive nutrient source for microorganisms, but still, trees can live for sometimes hundreds or even thousands of years. When a tree is attacked by animals or microbes it cannot run away, so it has to protect itself by other means, one of these being the outer defensive shell – the bark (Fig. 1). Similar to animal skins or exoskeletons, the bark can gradually peel off and thereby remove various parasites, but a probably more potent defense mechanism is that the bark differs chemically from wood by being enriched in so-called extractive molecules, many of which are toxic or inhibitory to attackers. Despite this potent defense, once a tree has fallen the bark does not accumulate but is instead over time decomposed.Curiously, despite the bark being an easily recognized material by most people, very little is known about how microorganisms can actually tackle this material – which species drive the process? In which order are the bark components degraded, and especially, what happens to the toxic extractives? Previous studies of bark have mainly focused on determining its chemical composition, and there are a few studies which have looked into microbes growing on it, but in these either the bark chemistry perspective is missing or the temporal perspective, or both.

We started our biological bark degradation studies back in 2019, focusing mainly on spruce bark as spruce is a major species in the forestry industry in the Nordic region. We obtained bark from a mill in the middle of Sweden, where the general process is to strip tree logs from bark once they arrive at the mill and then collect the bark in huge piles before burning it. After milling the bark to a manageable size, we decided to gamma-irradiate it instead of using typical autoclaving for sterilization, to keep the material as untouched as possible. The idea to study bark degradation by microbial consortia was initially a side project for Amanda Sörensen Ristinmaa, the PhD student who has spent the last four years on this topic. Her main project focused instead on following degradation by individual microbial species, but due to various complications during the pandemic this was partly put on ice. For the community studies, we did not know if we could even extract good quality DNA from the complex bark material, but we reached out to the company DNASense who did some preliminary tests that worked very well. We then inoculated lots of sterile bark samples with a microbial consortium sourced from the original bark pile, one for each replicate and sampling point to avoid risks of contamination. The degradation was expected to be quite slow so Amanda set up enough samples to be able to follow the process for six months.
For each sampling point, samples were then collected and a part of it analyzed chemically to monitor carbohydrates, lignin and extractives, while the other part was sent for DNA extraction and sequencing. This enabled mapping both of what was going on in the material itself and also which types of species were present at the same time. Only a few samples had to be discarded along the way due to contamination, and overall it all worked surprisingly well, but of course also comprised a lot of analytical work. When combining the chemical and sequencing results, we could see that especially in the initial degradation the microbial diversity dropped which indicates that something in the bark either is inhibitory to many species or that only a selection of species can grow on fresh bark. We could link the recovery of the diversity with metabolism/modification of extractive molecules known as resin acids, and surprisingly there was little changes to the carbohydrates that we had expected to be a major food source for the microbes. We then chose to dig deeper into this initial degradation using metagenomics, and could see a correlation of genes linked to resin acid-degrading capabilities.
In parallel, Amanda set up some culture plates with resin acids as the sole carbon source and was able to isolate several bacterial strains. One was picked for sequencing and it turned out to be the most abundant bacterial strain in the consortium and also to represent a new species. Species isolation and naming is not what we have typically done in the group previously, and limited knowledge in Latin made it a bit of a challenge but eventually we came up with Pseudomonas abieticivorans, after also having considered P. abietanovorans, and apparently also the lab working name P. barky.
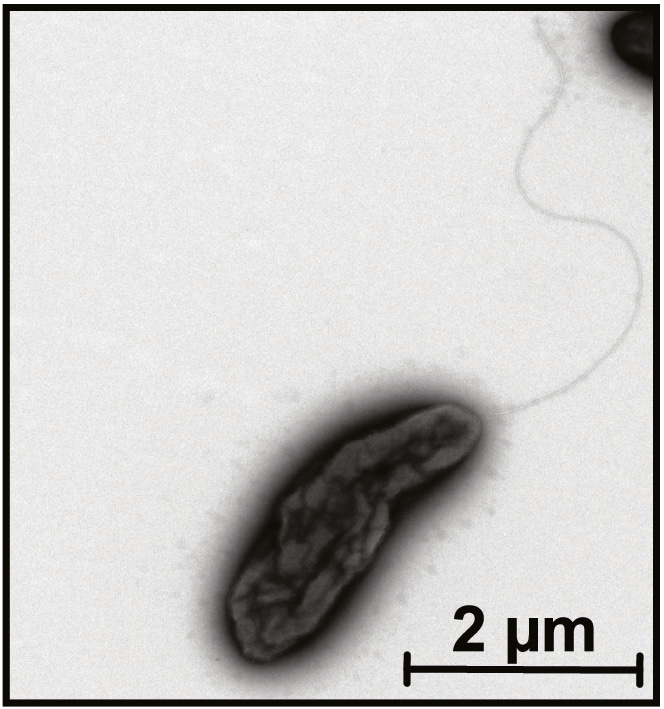
What does this study mean for degradation of spruce bark in nature? We believe that the resin acids are serving as gatekeeper molecules to keep most attackers at bay and only a few species are able to actively metabolize them, but possibly other species may be able to partially modify them to reduce their toxicity. Even in our conditions, the degradation was very slow, and this is seen also in forests where fallen trees (and their bark) are decomposed over many years. In a living tree, the resin acids would, in contrast to our conditions, be continuously supplied to the bark to further increase its resistance to attack and. The research can also be relevant for industry, as bark is a huge resource, produced in millions of tons annually, but it has a very low value. Most of the bark is burnt for energy, but whether this is a good use could be questioned as it is also a quite wet material, and enriched in minerals that leads to formation of more ash compared to burning of wood. Knowing how the bark can be degraded by microorganisms could open up new ways of using this structurally rich material, and also mitigate the losses to unwanted microbial degradation.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in