Modeling t(4;11) leukemia in umbilical cord hematopoietic stem precursor cells
Published in Biomedical Research
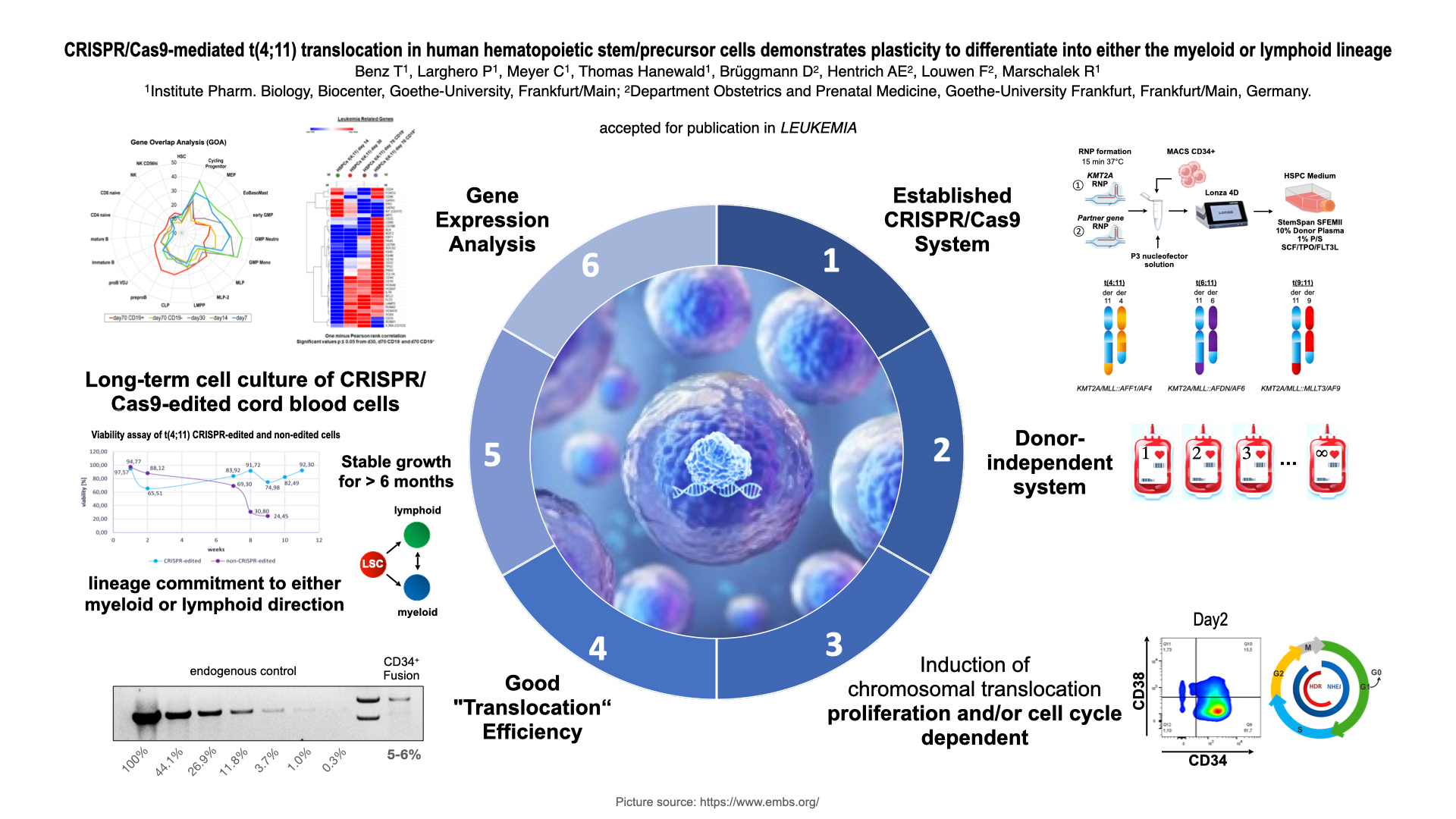
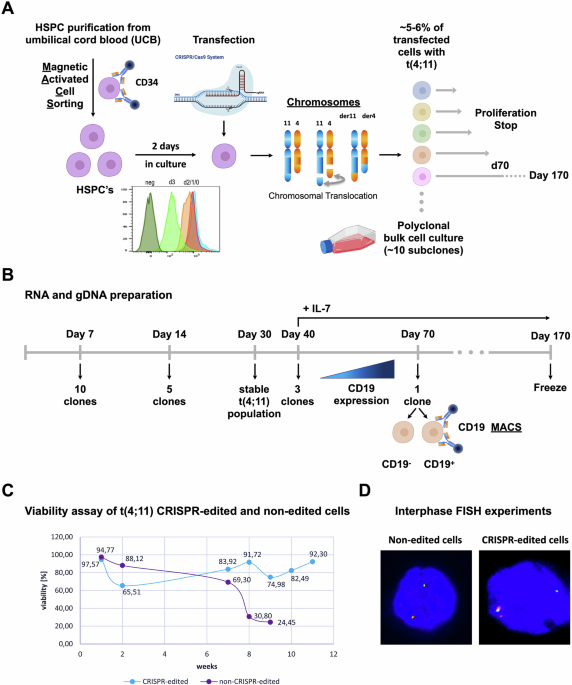
Chromosomal translocations of the KMT2A/MLL gene are frequently diagnosed in acute leukemia patients (ALL and AML) with very poor outcome. Although we have animal models for some of the frequent KMT2A/MLL fusions, a reliable model for the most frequent fusion – the t(4;11)(q21;23) translocation - is still missing. Experiments in animals by using the fusion genes KMT2A::AFF1 and/or AFF1::KMT2A were difficult to establish or have even failed. Therefore, we decided to model this translocation t(4;11)(q21;q23) in human cells by using the CRISPR/Cas9 technology in combination with CD34+ hematopoietic stem/precursor cells that were purified from umbilical cord blood. The reason for this effort is clear: although we understand much about the function of both wildtype KMT2A and AFF1 multiprotein complexes, very little is known about the molecular actions the two fusion proteins KMT2A::AFF1 and AFF1::KMT2A, in particular their precise function during the very early steps of disease onset and progression.
Here, we present the results of our efforts to establish a t(4;11) chromosomal translocation in human hematopoietic stem/precursor cells by CRISPR/Cas9. These genetically modified cells can be expanded over 5-6 months in vitro and their potential to differentiate was examined with IL-7 supplementation. Upon administration of IL-7, a small portion of cells (~5%) responded and differentiated in vitro into CD19+ pre-leukemic cells. Gene expression profiling revealed an 80% concordance with gene expression profiles derived from infant and non-infant t(4;11) patient cells. This indicates that we were able to model the t(4;11) leukemia disease ex vivo without the need of animal experiments. The benefit of this CRISPR/Cas9 model system is that (1) both reciprocal fusion proteins are concomitantly present, (2) a molecular surveillance is possible at any timepoint through analysis of RNA, DNA or protein, and (3) that this universal system can be used to model any other KMT2A rearrangement. Up to now, we have done so to model the translocation t(9,11)(p22;q23) (aka KMT2A::MLLT3 / MLLT3::KMT2A) and t(6;11)(q27;q23) (aka KMT2A::AFDN / AFDN::KMT2A). Currently, we are performing experiments to additionally model two t(11;19) translocations (aka KMT2A::ENL and KMT2A::ELL).
In conclusion, this experimental set-up - or similar approaches using fetal liver cells instead of umbilical cord blood cells - is the fastest way to investigate and characterize KMT2A-fusions in primary human cells. The possibility to transplant these cells directly into NOD/SCID mice to monitor engraftment and kinetics of leukemia development, or to culture these cells ex vivo over long time periods will help to understand the malignant conversion of normal hematopoietic stem/precursor cells into pre-leukemia and/or leukemic cells. These insights will help to identify critical steps and new venues for the treatment of these high-risk acute leukemia patients in the future.
The paper can be found in open access here: https://www.nature.com/articles/s41375-025-02791-4
Follow the Topic
-
Leukemia

This journal publishes high quality, peer reviewed research that covers all aspects of the research and treatment of leukemia and allied diseases. Topics of interest include oncogenes, growth factors, stem cells, leukemia genomics, cell cycle, signal transduction and molecular targets for therapy.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
https://link.springer.com/article/10.1007/s12552-025-09459-9
Chandra, B., Parasar, A. The Current Situation of People Belonging to the Dalit Community Living in Bangladesh to Achieve the 11th Goal of SDG 2030. Race Soc Probl (2025). https://doi.org/10.1007/s12552-025-09459-9