Molecular Analysis of Pseudomonas aeruginosa Isolates with Mutant gyrA Gene and Development of a New Ciprofloxacin Derivative for Antimicrobial Therapy
Published in Bioengineering & Biotechnology, Microbiology, and Cell & Molecular Biology

Study Background:
The primary focus of the research is to investigate the prevalence of Pseudomonas aeruginosa in various medical specimens. The study encompasses a comprehensive examination of 225 specimens from individuals aged 30 to 60, covering diverse sources such as wounds, burns, urine, sputum, and ear samples.
Genetic Analysis and Antibiotic Resistance:
The study utilizes advanced techniques like PCR–RFLP and gene expression analysis to identify resistant strains of P. aeruginosa and explore the genetic basis of antibiotic resistance, particularly in connection with the gyrA gene. Notably, the investigation establishes a correlation between ciprofloxacin resistance and mutant gyrA gene presence.
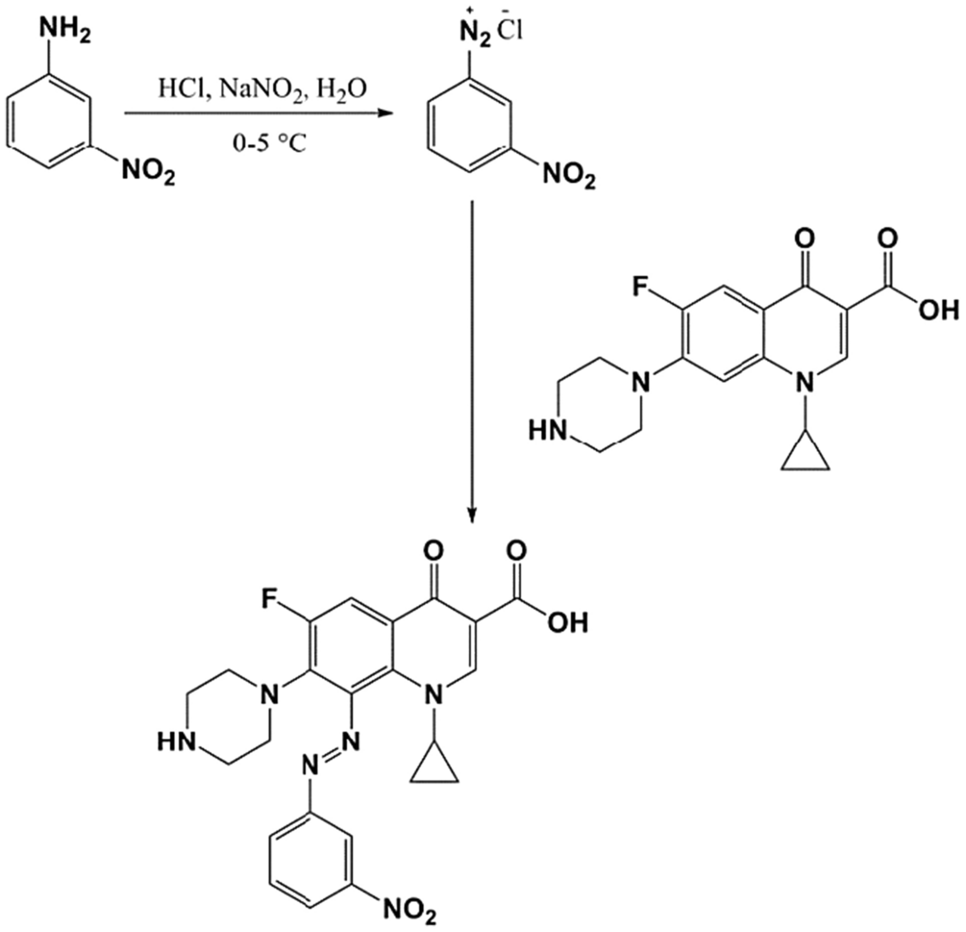
Synthesis and Testing of Ciprofloxacin Derivative:
A new ciprofloxacin derivative is synthesized and characterized using FT-IR, 1H-NMR, and mass spectroscopy techniques. The antibacterial efficacy of the derivative is tested, revealing promising results with MIC values ranging from 1 to 2 μg/ml.
Molecular Docking Study:
The research employs molecular docking techniques to predict the mechanism of action for the synthesized ciprofloxacin derivative. The findings illustrate the strategic positioning of the derivative within the DNA-binding site of the gyrA enzyme, indicating its potential as a novel antimicrobial agent.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in