New Tools for Screening for Enivronmentally Influenced Epigenetic Disease Risk Factors
Published in Cell & Molecular Biology and Genetics & Genomics
Background
Over the past 100 years, the primary causes of death in the developed world have shifted from infectious diseases to non-infectious diseases; in the United States in 1900, influenza and tuberculosis were the top two causes of death, whereas in 2018, heart disease and cancer were the top two causes of death. In part, these shifts are due to the reduction of infectious diseases by improved medical technology, so that a larger proportion of the population is living long enough to develop age-related diseases. However, demographic shifts towards an older population do not fully account for increased incidence of some diseases, such as metabolic syndrome, liver disease, and certain cancers. These diseases have had dramatic increases in the percentage of people affected over the last 30 years. There are also diseases with disparities depending on sex and ethnic background, with differences in age of onset, rate of progression, and overall severity.
While there are genetic and environmental factors associated as risks for these diseases, they are not definitive that the disease will occur, and are only found in a subset of affected people. A better understanding of what the risks are, and how genes and environment (nature and nurture) interact to cause disease, will give us new tools for early detection and intervention, and new possibilities for treatment.
Epigenetics
Epigenetic mechanisms control how DNA is packaged, by directly methylating DNA bases and modifying histone proteins that DNA wraps around, with the effect of regulating gene expression. Epigenetic factors have an essential role in embryonic development, by guiding the specialization of cell types, and regulating the correct growth of tissues and organs. However, this epigenetic regulation is sensitive to environmental exposures, and can change how an animal or person grows, based on the environment they live in. Because epigenetic marks are actual chemical changes to DNA and proteins, they can be long-term recordings of what someone has lived through, and even be passed on to their children. Like an inherited mutation, inherited epigenetic marks can create changes in offspring, but unlike mutations that that change the actual DNA sequence, epigenetic marks are flexible, and can be created and removed, so that their effects can be created and reversed. Therefore, determining how specific genes are controlled by epigenetic marks, and how the environment can affect these epigenetic marks, offers possibilities to understanding what raises and lowers the risks of complex non-contagious diseases, and how exposures from the modern world and lifestyle can be increasing the incidence of these diseases (Figure 1).
Figure 1 – Epigenetics as a mechanism for translating environmental exposures to outcomes.
Imprinting and Imprint Control Regions
We are particularly interested in the set of epigenetic markers that make up “Imprint Control Regions” (ICRs), which regulate “Imprinted genes” – genes which are expressed from only one of the two allelic copies. ICRs are methylated on only one allele, determined by which parent the allele was inherited from, subsequently regulating the activation/silencing of the two alleles of target genes based on the parent of origin (Figure 2). Imprinted genes are particularly important in development, as most of the known imprinted genes are developmental regulatory genes, with the set of genes expressed from the father’s copies tending to promote growth, and the set of genes expressed from the mother’s copies tending to restrict growth. Keeping this imprinting under strict control is important because these genes are developmental regulators that must have specific expression amounts for proper growth. Developmental disorders such as Prader-Willi and Angelman syndromes are due to complete loss of control of specific imprinted genes, and it is suspected that slight alterations in imprinted gene expression affect the development of disorders later in life. ICRs are also a potentially powerful diagnostic tool, as their methylation is established at the single-cell stage of development, and maintained consistently in all subsequent cell types. Therefore, any perturbations to ICR methylation that happen in development should be detectable in blood DNA, as a proxy for cell types inaccessible for screening in people with no diagnosed illness (i.e. brain, liver, heart).
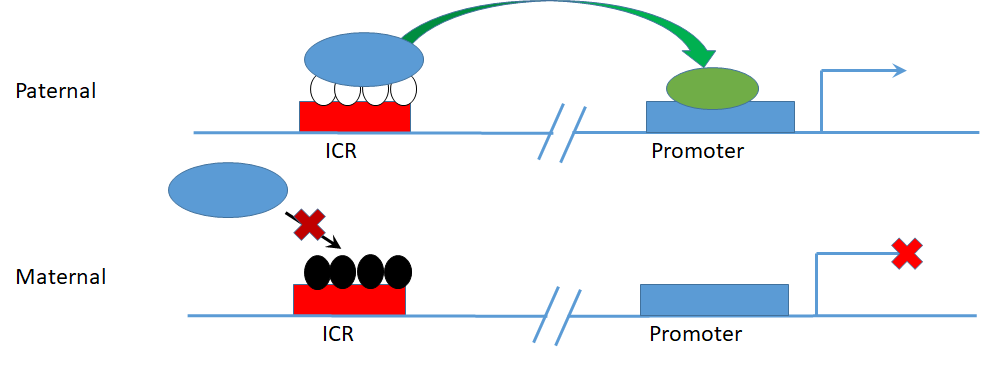
Figure 2 – Parent of origin specific allele methylation at an ICR regulating imprinted allele specific gene expression.
Using whole genome sequencing methods to quantitate methylation, we have identified 1488 regions in the human genome that have methylation patterns consistent with what an ICR is expected to look like (1)(Figure 2). With these potential ICRs identified, our goal became to identify how the methylation is altered in complex diseases, and in association with specific environmental exposures. This requires the screening of large case-control sets, in order to make definitive decisions about which ICRs are significantly affected. The problem is, that sequencing the number of samples (hundreds) required is not time or cost effective, and also requires large amounts of data storage and processing. Additionally, once the aberrantly methylated regions are identified, and we wish to put this information into use in diagnostic screening, which needs a straightforward method that can be applied and analyzed in a clinical setting.
Custom ICR Methylation Array
To make ICR screening more accessible and economical, we partnered with TruDiagnostic to design a custom Illumina methylation array, targeting the set of human ICRs, that can be broadly implemented for identifying aberrant methylation, and early detection of disease risk. The array generated can measure methylation at over 22,000 DNA sites, covering 1088 (74%) of the 1488 ICRs we have identified, in comparison to the ~10% of ICRs covered by previous methylation arrays. Using this array on a case-control set of Alzheimer samples, using both autopsy brain tissue, and blood from living patients, we have determined that the array methylation measurements correlate well to the whole genome methylation sequencing. The ICR methylation measured by the arrays is highly replicable upon repeated sample analysis, and showed the expected intermediate level of methylation expected for ICRs with monoalleleic methylation (2). These arrays are now being implemented in case-control studies to identify ICRs with altered methylation, pointing the way to new diagnostic tools, and the underlying disease mechanisms that can be targets for treatment.
References
- Jima DD, Skaar DA, Planchart A, Motsinger-Reif A, Cevik SE, Park SS, Cowley M, Wright F, House J, Liu A, Jirtle RL, Hoyo C. Genomic map of candidate human imprint control regions: the imprintome. Epigenetics. 2022 Dec;17(13):1920-1943. doi: 10.1080/15592294.2022.2091815. Epub 2022 Jul 4. PMID: 35786392; PMCID: PMC9665137.
- Carreras-Gallo N, Dwaraka VB, Jima DD, Skaar DA, Mendez TL, Planchart A, Zhou W, Jirtle RL, Smith R, Hoyo C. Creation and Validation of the First Infinium DNA Methylation Array for the Human Imprintome. bioRxiv [Preprint]. 2024 Jan 16:2024.01.15.575646. doi: 10.1101/2024.01.15.575646. PMID: 38293193; PMCID: PMC10827131.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in