Pan-Cancer Analysis of PTPN6: Prognostic Significance and Functional Implications in Tumor Progression
Published in Cancer and Biomedical Research
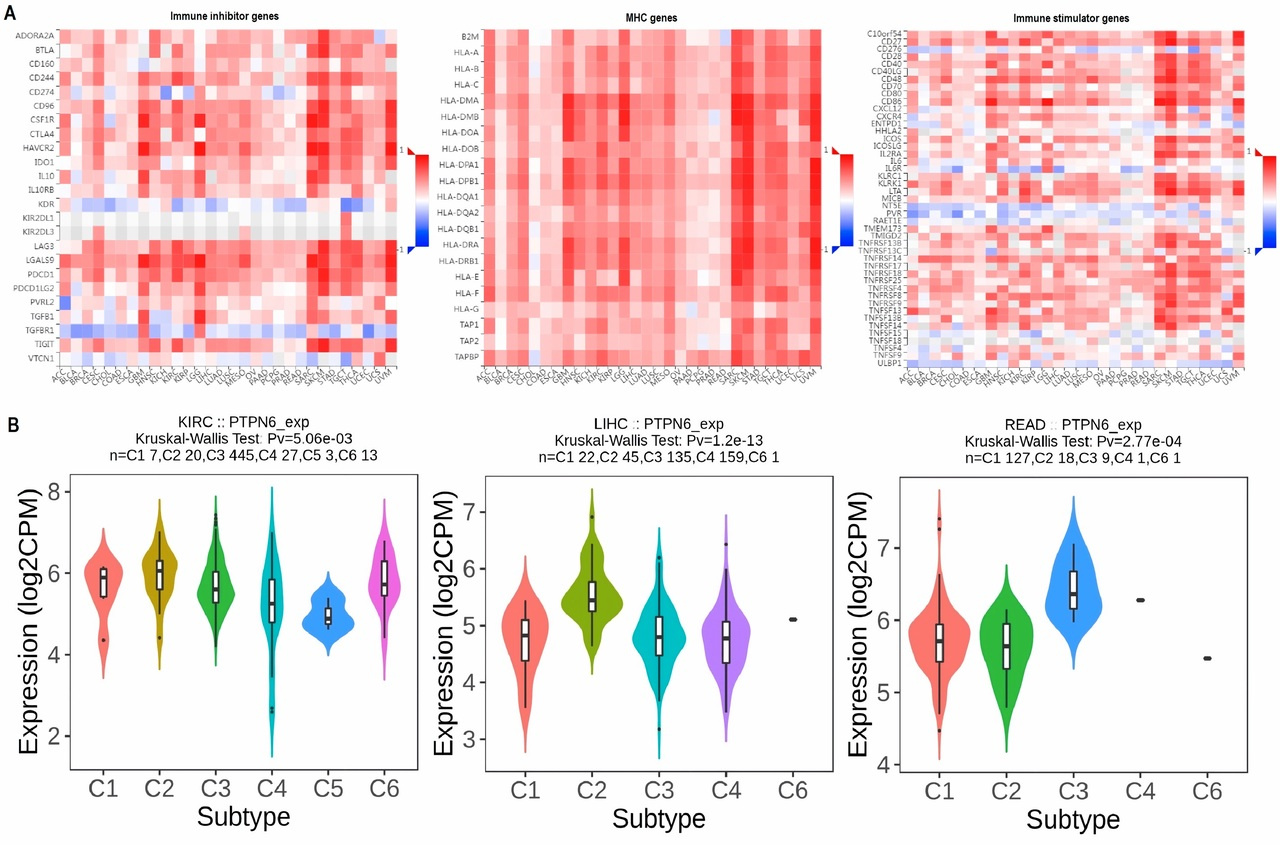
This paper aimed to address that gap, focusing on PTPN6, a gene known for regulating immune responses and cell signaling, which had already been studied in blood cancers like lymphoma and leukemia. However, its role in other cancer types, especially solid tumors, wasn’t well understood.
The research team, a group of scientists from different countries and fields, wanted to see if PTPN6 could be a common factor across many cancers, offering new ways to diagnose or treat them. While PTPN6 had shown some promise in earlier studies, it hadn't been explored much across a wide range of cancers. So, the team started by analyzing data from several public databases, hoping to get a clearer picture of how PTPN6 behaved across different cancer types. The results were striking. They found that PTPN6 expression was different between cancerous and normal tissues, which made it a potential biomarker. More importantly, they discovered that higher levels of PTPN6 were linked to worse survival outcomes in cancers like kidney renal clear cell carcinoma (KIRC), liver hepatocellular carcinoma (LIHC), and rectum adenocarcinoma (READ).
What the team found next raised more questions. The role of PTPN6 appeared to depend on the context of the cancer. In some cancers, it seemed to act like a tumor suppressor, while in others, it might be contributing to tumor growth. It was clear that the relationship between PTPN6 and cancer wasn’t simple. Further analysis showed that changes in PTPN6 expression were tied to DNA methylation changes, hinting that there was more to its regulation than previously thought. The team also ran functional tests on cancer cell lines. When they manipulated PTPN6 levels in the lab, the cells changed their behavior, growing and migrating more quickly, which are key traits in cancer progression.
The researchers also looked at the mutations in PTPN6 across different cancers. While mutations were rare, the upregulation of PTPN6 in certain cancers seemed to be a consistent feature. This led the team to ask whether the gene’s expression was causing cancer progression, or if it was part of a broader mechanism affecting the tumor’s environment.
At this point, the team felt confident that PTPN6 was a promising biomarker and a potential target for treatment. They knew that their results could be used to develop more precise cancer therapies, especially for cancers where PTPN6 was overexpressed. While the findings showed that PTPN6 might be important for prognosis, the exact way it contributes to cancer progression still needed further research.
Despite the complexity of the project and the technical challenges, the team was motivated by the potential impact of their work. They knew that their study could lead to a better understanding of how cancer behaves at the molecular level and, ultimately, help improve treatment options for patients. Looking back, they were proud of how their collaboration led to this paper, which could now serve as a foundation for future studies. Though the journey to fully understanding PTPN6’s role in cancer is far from over, the team had made an important contribution to cancer research, and they hoped that their findings would guide the way for new, targeted therapies.
Follow the Topic
-
Discover Oncology

This is a fully open access general oncology journal that aims to provide a unified forum for researchers and clinicians. The journal spans from basic and translational science, to preclinical, clinical, and epidemiology, and welcomes content that interfaces at all levels of cancer research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Single-Cell RNA Sequencing in Cancer Immunotherapy
Cancer immunotherapy is a hot area of current oncology research, with its core focus on activating or enhancing the body's immune system's ability to recognize and kill cancer cells. However, cancer cells possess complex heterogeneity and dynamics, which affect the efficacy of immunotherapy in many ways. Single-cell RNA sequencing (scRNA-seq) has emerged as a powerful tool in recent years, providing us with an unprecedented insight into the cellular heterogeneity and dynamics within tumors. This technology has revolutionized our understanding of cancer biology, especially in the context of cancer immunotherapy. By enabling researchers to analyze individual cells, scRNA-seq allows them to identify distinct cell populations, track cellular responses to treatments, and discover new therapeutic targets. This collection aims to compile cutting-edge research in this field and explore the various applications of single-cell RNA sequencing in cancer immunotherapy.
This collection will cover the following topics: 1. The latest advances in single-cell RNA sequencing technology in cancer immunotherapy, including research on technology optimization and data interpretation; 2. Using single-cell RNA sequencing to reveal the characteristics of immune cell subgroups in the tumor microenvironment and their interaction mechanisms with cancer cells; 3. Analyzing the molecular basis of immune therapy response and resistance through single-cell RNA sequencing, exploring new biomarkers and therapeutic targets; 4. Combining single-cell RNA sequencing with clinical studies of immunotherapy to assess treatment outcomes, predict patient prognosis, and optimize treatment plans.
Keywords: cancer immunotherapy, single-cell RNA sequencing, therapeutic targets, tumor microenvironment, treatment response
Publishing Model: Open Access
Deadline: Jun 30, 2026
Tumor Microenvironment
The immunological and stromal components of the tumor microenvironment (TME) play important roles in supporting or preventing tumor growth. The interaction of tumor cells and immune cells inside the TME is complex, and it can result in immune suppression, immune evasion, or, in the opposite case, successful immune-mediated tumor elimination. Cancer therapy has increasingly focused on targeting the TME. Immune checkpoint inhibitors (e.g., anti-PD1, anti-CTLA4), CART cell therapy, and vaccinations are all aimed at reactivating the immune system's ability to recognize and destroy tumor cells. Modulating immune component activity is being investigated as a way to counteract the immunosuppressive TME and boost immunotherapy efficacy. This thematic Collection will provide a thorough understanding of the complex role of the tumor microenvironment in the emergence of cancer treatment resistance, as well as innovative strategies for overcoming this challenge and investigating the tumor microenvironment's contribution to personalized medicine.
Keywords: tumor cells, TAMs, TILs, stromal cells, immunotherapy
Publishing Model: Open Access
Deadline: Jun 30, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in