
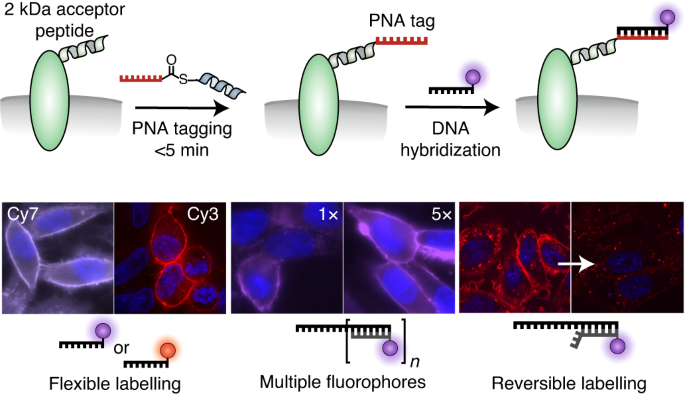
The clichéd ‘lightbulb moment’ that eventually lead us to our paper PNA tagging – A Method for Versatile and Erasable Fluorescence Imaging of Membrane Proteins took place at the 13th Akabori Conference in Leipzig in 2010, where Katsumi Matsuzaki (Kyoto University, Japan) presented his research on coiled coil interactions for non-covalent labelling of membrane proteins. When Oliver heard the talk, he immediately realised that he could use the same coiled coils to enable covalent labelling. Our group had already used PNA (peptide nucleic acid- essentially DNA with a peptide backbone) conjugates to achieve templated reactions, in which a reporter group or a peptide was transferred from a thioester linked PNA to a Cysteine-PNA. Merging this with Matzuzaki’s labelling would just require switching the PNA for the coiled coil peptide.
Fast forward 10 years and the idea has developed into a live cell fluorescent labelling technique which encompasses many aspects one strives for in a biorthogonal labelling reaction; namely specificity, speed, small tag size, versatility and reversibility. Back to the time when everything started, the German research funding organisation Deutsche Forschungsgemeinschaft established a nation-wide priority program named "Chemoselective reactions for the synthesis and application of functional proteins" which allowed Oliver to team up with the group of Annette Beck-Sickinger (Universität Leipzig, Germany) to realise the labelling idea in the context of living cells. The coiled coil-templated transfer of a fluorophore reporter worked beautifully and enabled rapid labelling of membrane proteins on live cells. Publications followed, with the Beck-Sickinger group using the technique to analyse rapid signalling patterns of GPCRs.
Meanwhile in the Seitz group Katherina Gröger was further developing the technique to transfer a PNA strand rather than a fluorophore. This made sense in the context of our group; we have a longstanding interest in PNA, and oligonucleotide conjugates were used as a tool for a variety of applications such as imaging mRNA or DNA programmed spatial screening of multivalent protein binding sites. Given the knowledge of what oligonucleotide arrangement can achieve in a biological setting, using PNA as a protein tag seemed only natural.
And so the idea seemed to have come in a full circle to unite peptide and oligo chemistries once again: from PNA-templated transfer of peptide, to peptide-templated transfer of PNA. This is about the time when I showed up. Working in such a group where people are comfortably switching from peptides to DNA, organic synthesis to cell culture and microscopy, there was certainly a lot to learn. But from this project I see how working on the boundary of biology and chemistry, with DNA and peptides can be beneficial. Aside from the fact that there’s something amazing about synthesising a molecule from scratch and taking it all the way to live cell labelling reactions; being involved in all these circles helps bring together ideas from different disciplines. It also helps if you have real biologists to step in when things get serious (i.e. our wonderful collaborators Michael Bartoschek from the Bultmann group at LMU Munich, and Philipp Wolf from the Beck-Sickinger group).
Once the PNA transfer reactions worked, it was just a matter of following the literature. Toehold-mediated strand displacement introduced by Yurke worked amazingly to allow erasable labelling. But not only can one imagine the PNA tag used for all sorts of other clever DNA technologies (DNA-PAINT, spatial organisation, proximity ligation etc.) but using a rationally designed coiled coil system as the basis of target recognition will also enable further developments- the Jerala group have already shown that a set of 6 orthogonal coiled coils can be functional in a cellular setting. Although I won’t be involved in all of this before my time finishes in the group, I feel at least contented that the next lucky PhD candidate will have their hands pretty full.
Our paper can be found at: https://www.nature.com/articles/s41557-020-00584-z
Further Reading:
“Coiled-coil tag--probe system for quick labeling of membrane receptors in living cell. “ Yano Y, Yano A, Oishi S, Sugimoto Y, Tsujimoto G, Fujii N, Matsuzaki K. ACS Chem Biol. 2008 Jun 20;3(6):341-5. doi: 10.1021/cb8000556. PMID: 18533657.
"Rapid Covalent Fluorescence Labeling of Membrane Proteins on Live Cells via Coiled-Coil Templated Acyl Transfer" U. Reinhardt, J. Lotze, K. Mörl, A.G. Beck-Sickinger, O. Seitz*, Bioconjugate Chem. 2015, 26, 2106-2117 (DOI: 10.1021/acs.bioconjchem.5b00387)
”Time-Resolved Tracking of Separately Internalized Neuropeptide Y2 Receptors by Two-Color Pulse-Chase" J. Lotze, P. Wolf, U. Reinhardt, O. Seitz, K. Mörl, A.G. Beck-Sickinger*, ACS Chem. Biol. 2018, 13, 618–627 (DOI: 10.1021/acschembio.7b00999)
“A tunable orthogonal coiled-coil interaction toolbox for engineering mammalian cells. Nature Chemical Biology” Lebar, T., Lainšček, D., Merljak, E., Aupič, J. & Jerala, R. Nat Chem Biol 16, 513–519 (2020).doi:10.1038/s41589-019-0443-y (2020).
“A DNA-fuelled molecular machine made of DNA.” Yurke, B., Turberfield, A., Mills, A. et al. Nature 406, 605–608 (2000). https://doi.org/10.1038/35020524
Follow the Topic
-
Nature Chemistry

A monthly journal dedicated to publishing high-quality papers that describe the most significant and cutting-edge research in all areas of chemistry, reflecting the traditional core subjects of analytical, inorganic, organic and physical chemistry.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in