Pre-training a Foundation Model for Generalizable Fluorescence Microscopy-Based Image Restoration
Published in Protocols & Methods

Fluorescence microscopy is an indispensable tool for studying the spatiotemporal dynamics of cell and tissue development in life sciences, enabling high-resolution imaging of biological processes within cells. However, its imaging quality is limited by the microscope optics and the achievable fluorophore density, bleaching, and photo-toxicity1.
Fluorescence microscopy-based image restoration has received widespread attention in the life sciences for its ability to break through the physical limits of fluorescence microscopy imaging, achieving better imaging speed, spatial resolution and depth. Benefiting from deep learning technology, fluorescence microscopy-based image restoration1,2,3,4 has led to significant progress, better revealing the complex interactions and dynamics inside cells and biomolecules, and expanding the scope of observation of biological phenomena. However, most current task-specific methods have limited generalizability to different fluorescence microscopy-based image restoration problems.
Here, we seek to improve generalizability and explore the potential of applying a pre-trained foundation model to fluorescence microscopy-based image restoration. We provide a unified foundation model (UniFMIR) to address different restoration problems, including super-resolution, 3D denoising, isotropic reconstruction, image projection and volumetric reconstruction (Figure 1), and show that UniFMIR offers higher image restoration precision, better generalization, and increased versatility.
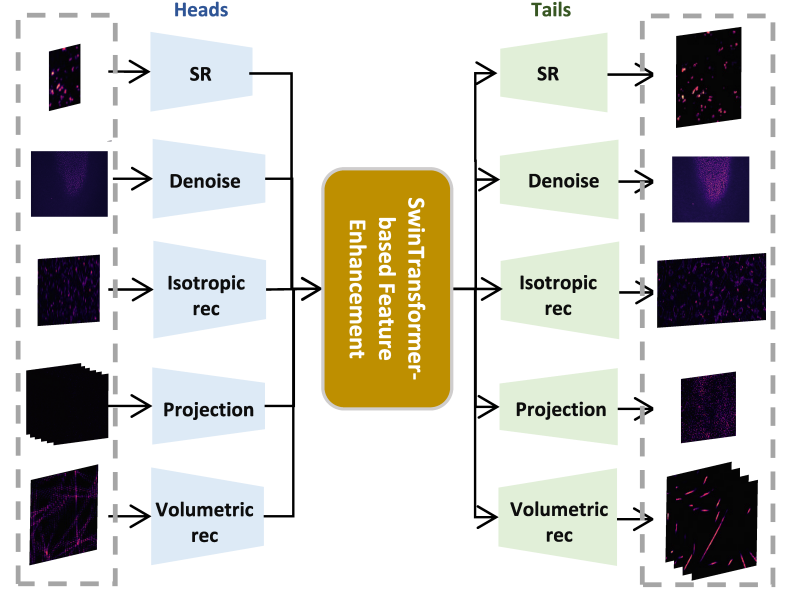
Figure 1. Architecture of the proposed UniFMIR, which consists of three components, including multiple feature extraction modules, a Swin transformer-based feature enhancement module, and multiple image reconstruction modules. The multi-head and multi-tail branches of different network structures are adopted to extract task-specific shallow features and reconstruct images for different fluorescence microscopy-based image restoration tasks.
To demonstrate the generalization ability of our pretrained UniFMIR approach, we validated its image restoration performance on unseen data from DeepBacs4 for bacterial image analysis purposes, including two denoising datasets (E. coli_H-NS-mScarlet-I, E. coli_MreB) and two superresolution datasets (E. coli, S. aureus). We fine-tuned UniFMIR on the new bacterial microscopy images and then conducted denoising and superresolution experiments. In addition to the denoising and superresolution models developed on DeepBacs, we also compared our results with those of a baseline model trained from scratch, which had the same structure as UniFMIR, and the pretrained UniFMIR model without fine-tuning. Our UniFMIR method restored clear prokaryote structures to enhance the low-phototoxicity live-cell microscopy data and predict accurate mappings of biological target shapes, obtaining higher PSNR/SSIM values (Figure 2).
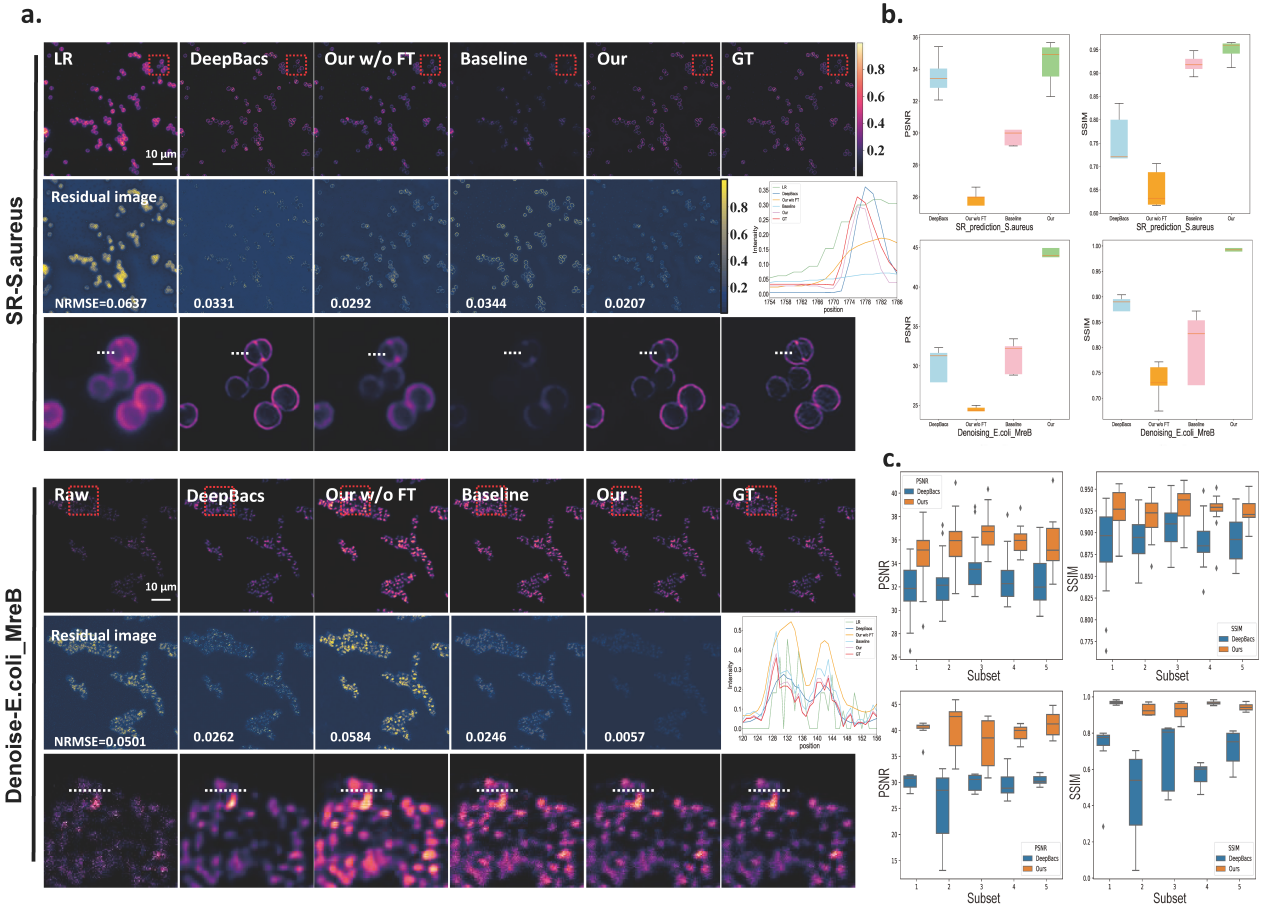
Figure 2. Generalization ability analysis conducted on unseen datasets4. a, SR and denoising results obtained on the S. aureus and E. coli_MreB datasets, respectively. Visual comparison among the outputs of the SOTA model (DeepBacs4), pretrained UniFMIR model without fine-tuning, Baseline (same network structure as UniFMIR trained from scratch), and our fine-tuned UniFMIR model. The NRMSE is shown on each residual image. b, The PSNR/SSIM results obtained on the test sets of the two datasets. c, K-fold validation (K=5) on the S. aureus dataset (up) and the E. coli_MreB dataset (down). The PSNR/SSIM results were obtained on the n=5 test images of different subsets. Scale bar: 10 µm.
UniFMIR provides a universal solution for fluorescence microscopy-based image restoration. Pre-trained UniFMIR can be applied to different tasks, imaging modalities and biological structures through simple fine-tuning, demonstrating the great role of foundation model methods in biological imaging research. We believe that the image restoration capabilities of UniFMIR can be continuously enhanced by further expanding the amount and diversity of training data.
- Weigert, M. et al. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nature methods 15, 1090-1097 (2018).
- Qiao, C. et al.Evaluation and development of deep neural networks for image super-resolution in optical microscopy. Nature methods, 1–9 (2021).
- Wang, Z. et al. Real-time volumetric reconstruction of biological dynamics with light-field microscopy and deep learning. Nature Methods 18, 551–556 (2021).
- Spahn, C. et al. Deepbacs for multi-task bacterial image analysis using open-source deep learning approaches. Commun Biol 5, (2022).
Follow the Topic
-
Nature Methods

This journal is a forum for the publication of novel methods and significant improvements to tried-and-tested basic research techniques in the life sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Methods development in Cryo-ET and in situ structural determination
Publishing Model: Hybrid
Deadline: Jul 28, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in