Proteomic analysis of archival breast cancer clinical specimens identifies biological subtypes with distinct survival outcomes
Published in Cancer
Background:
Over the past 20 years, breast cancer has been recognized as a family of diseases rather than one disease, consisting of unique entities with distinct pathological, molecular, and clinical features.
The genomic subtyping of breast cancers into luminal A, luminal B, human epidermal growth factor receptor-2 (Her2)-Enriched and basal-like has remarkably advanced breast cancer diagnosis, treatment, and outcome1,2. Despite this, extensive heterogeneity still characterizes breast cancers beyond their DNA or RNA profiles especially within the aggressive basal-like subtype3. This group is clinically approximated as triple negative breast cancer, characterized by lack of immunohistochemical staining for estrogen receptor, progesterone receptor, and Her2.
Current clinical tests and treatment decisions are commonly based on protein level information and newer classifications based on proteomic profiling have been proposed during the past five years as a new promising approach to reveal the functional differences behind breast cancer heterogeneity and develop even more clinically-relevant classifiers in breast cancer. While these studies provided high-quality data to classify breast cancers4-6, they often had limited number of cases analyzed, lacked clinical outcomes, and required large amount of fresh-frozen tissue not typically available from patients, hindering their potential application in the clinical setting.
Why did we perform this study?
This is the fruit of a decade of diligent teamwork efforts where we aimed to develop a clinically-applicable classification of breast cancer using routine clinical specimens consisting of standard formalin-fixed paraffin-embedded (FFPE) because such specimens represent the most widely available materials in most bio-repositories, typically have associated clinical treatment and outcome data, and because this is the format routinely used for patient diagnosis. We hoped that our proteomic profiling will be specifically able to reveal the heterogeneity within basal-like and triple negative subtypes and to obtain information that can more easily be translated into clinical actionable tests.
What did we investigate?
We used our recently developed method by Hughes et al.7,8 termed Single-Pot, Solid-Phase-enhanced, Sample Preparation-Clinical Tissue Proteomics (SP3-CTP) which can capture biological features in FFPE tumor samples. Using the SP3-CTP method, we performed a broad scale global proteome profiling of a total 300 well-characterized archival FFPE breast cancer specimens, 75 of each of the four major PAM50 subtypes. These specimens were collected from patients diagnosed with invasive breast cancer in the periods of 2008-2013 (n=178) and 1986-1992 (n=122) across the province of British Columbia, Canada and we linked results with detailed clinical outcomes.
What did we report?
Our proteomic analysis quantified 9088 proteins in total and 4214 proteins across all 300 samples. Key results showed that within the PAM50-classified basal-like cases, proteomic profiling revealed two subgroups with one having “immune hot” expression features and highly favorable survival rates vs. another with “immune cold” expression features and poor clinical outcomes.
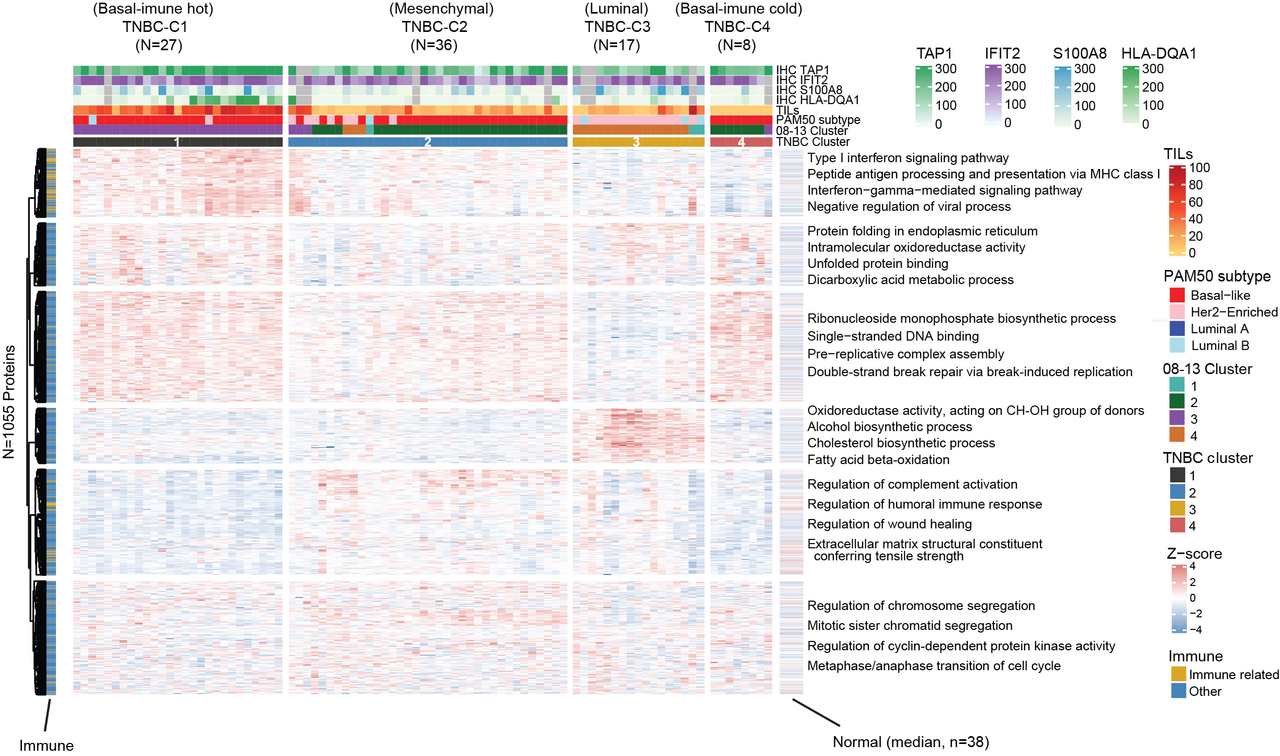
For triple negative breast cancers, which are of a particular clinical significance, our analysis of 88 clinical specimens identified four main proteomic subgroups that display protein features of the four most established RNA-based triple negative breast cancer subtypes9 of “basal-immune hot”, “basal-immune cold”, “mesenchymal”, and “luminal” and these displayed distinct survival outcomes (Fig. 1).
As part of biomarker development, we further verified and validated the expression of two biomarkers of TAP1 (MHC class I) and HLA-DQA1 (MHC class II) found to be characteristic of the proteomic immune hot group by immunohistochemistry. The combinatorial expression of these two immunohistochemical biomarkers was associated with the most favorable survival rates while the loss of these two biomarkers was associated with the worst survival on both discovery and validation sets.
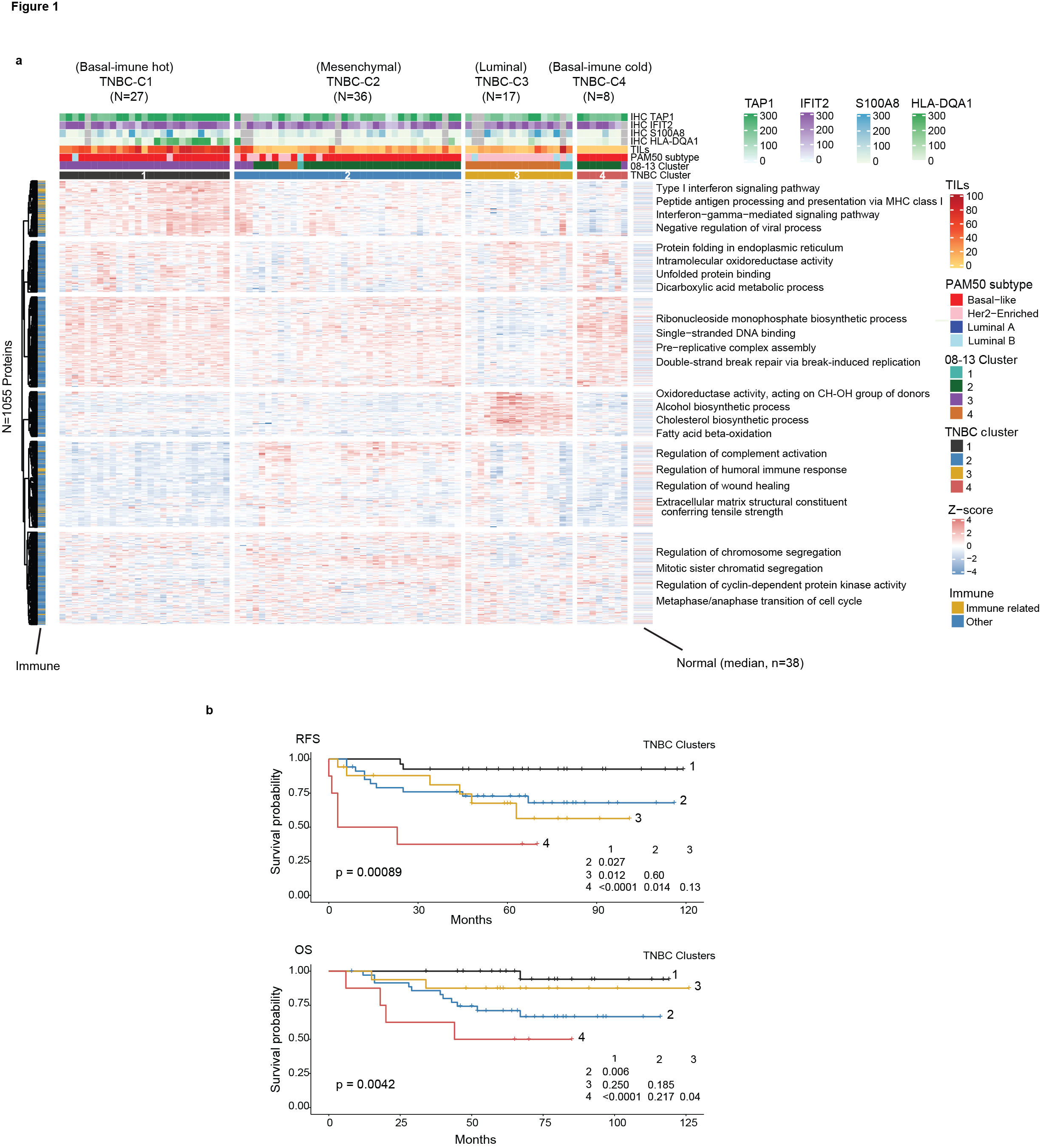
Fig.1 Proteomic analysis reveals four biologically and clinically distinct subtypes in triple negative breast cancer. (a) Consensus clustering of 88 immunohistochemistry-defined triple negative breast cancer cases reveals four subtypes based on the relative abundance of 1055 most variant proteins. (b) Kaplan Meier plots for recurrence free survival and overall survival across the triple negative breast cancer proteome clusters.
Why is our study impactful?
Our study demonstrates that a newer classification based on a comprehensive protein profiling expression using a method compatible with FFPE clinical specimens reveals the heterogeneity in breast cancer overall and specifically within the aggressive subtypes of basal-like and triple negative breast cancer. The link of our results to clinical outcome data characterizes this heterogeneity in a clinically-applicable manner, providing information that can be potentially translated into clinical tests.
What meaningful insights we bring into the most challenging forms of breast cancer?
Our study opens a new avenue to the field of triple negative and basal-like breast cancer subtypes, setting the ground for the development and validation of clinically-applicable proteomic classifier that tackles their diversity with a direct link to clinical outcome data. These subtypes account for approximately 15% of breast cancers overall with a propensity to afflict younger women, display the most aggressive clinical behaviour and have a higher rate of early recurrences when compared to other subtypes. The realization that these tumors are highly heterogenous presents the most pressing challenge of breast cancer translational research and highlights the need to develop biomarkers that can aid tailoring treatments for these subtypes in the clinical setting. Notably, the identification of a subgroup with favourable survival that shows high expression of immune response proteins can be used to identify potential biomarkers for existing chemotherapies, emerging immunotherapies or novel treatments that are much needed for these aggressive subtypes. Improving diagnostic tools and therapeutic options through developing better protein-based diagnostic tests using proteomics technologies are inherently advantageous in this regard.
Why our proteomic resource has a utility for biomarker discovery?
References
- Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. Aug 2000;406(6797):747-52. doi:10.1038/35021093
- Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. Mar 2009;27(8):1160-7. doi:10.1200/JCO.2008.18.1370
- Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. Oct 2012;490(7418):61-70. doi:10.1038/nature11412
- Mertins P, Mani DR, Ruggles KV, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 06 02 2016;534(7605):55-62. doi:10.1038/nature18003
- Krug K, Jaehnig EJ, Satpathy S, et al. Proteogenomic Landscape of Breast Cancer Tumorigenesis and Targeted Therapy. Cell. 11 25 2020;183(5):1436-1456.e31. doi:10.1016/j.cell.2020.10.036
- Johansson HJ, Socciarelli F, Vacanti NM, et al. Breast cancer quantitative proteome and proteogenomic landscape. Nat Commun. 04 08 2019;10(1):1600. doi:10.1038/s41467-019-09018-y
- Hughes CS, Foehr S, Garfield DA, Furlong EE, Steinmetz LM, Krijgsveld J. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol Syst Biol. Oct 2014;10:757.
- Hughes CS, McConechy MK, Cochrane DR, et al. Quantitative Profiling of Single Formalin Fixed Tumour Sections: proteomics for translational research. Sci Rep. 10 2016;6:34949. doi:10.1038/srep34949
- Burstein MD, Tsimelzon A, Poage GM, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. Apr 2015;21(7):1688-98. doi:10.1158/1078-0432.CCR-14-0432
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in