Reliability and accuracy of single-molecule FRET studies for characterization of structural dynamics and distances in proteins
Published in Protocols & Methods

More than 25 years after its inception [1], single-molecule FRET has matured into a routine tool to probe the mechanism and structure of macromolecules with applications in many fields ranging from structural biology over biochemistry to nanotechnology [2]. By coupling the structural information to a spectroscopic readout, smFRET experiments inform on distance changes in the range of 4-12 nm and are easily combined with single molecule observation. Notably, rather than providing snapshots of conformations, smFRET experiments offer correlated information on structure and dynamics on timescales ranging from nanoseconds to seconds/minutes.
In 2018, a benchmark study showed that FRET efficiencies could be measured with high reproducibility by 19 laboratories with subnanometer precision and accuracy [3]. This study emerged from a multi-year effort that was initiated by Thorsten Hugel in 2014 and coordinated by Björn Hellenkamp and Sonja Schmid. At that time, we were first year PhD students in the lab of Don Lamb at LMU Munich doing our first single-molecule FRET experiments. The task of participating in this study seemed daunting at that time – what if we turn out to be a clear outlier? This was amplified by the fact that we were not only building our microscopes from scratch but also writing our own analysis software (more on this below). As it turns out, however, we were not alone in this. Despite the wide variety of implementations of algorithms and setups, the field reached single-base pair resolution. Other valuable assets from the study were 1) a clear description and standardization of analysis routines, and 2) the founding of the "FRET community” which has hosted several workshops since then.
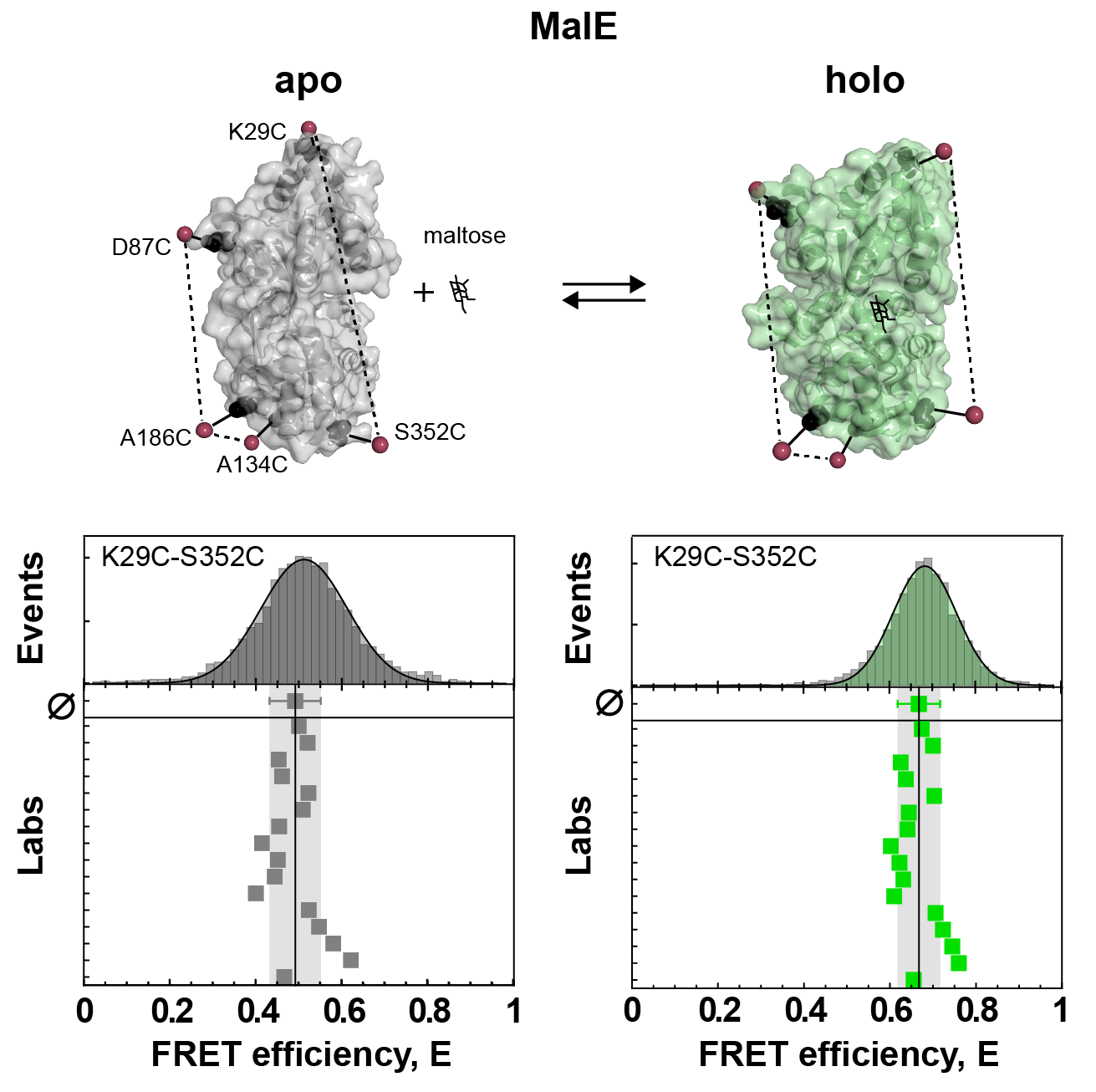
There were two limitations to this first “FRET challenge”. First, it was restricted to DNA molecules. Second, these DNA duplexes were essentially rigid rods. Hence, soon after, a second FRET challenge was initiated by Thorben Cordes, this time with a focus on proteins. This poses additional challenges for FRET experiments, as the biomolecular environment is much more diverse compared to DNA which can cause problems due to sticking or quenching of the fluorescent dyes. The first round focused on the widely-used maltose binding protein (MalE). MalE can be readily switched between open and closed conformation by the ligand maltose, which was reproducibly measured by the participating labs (see Figure 1 and below). This was a promising result, however it failed to address the topic of dynamics, because MalE assumed either the open apo state or the closed holo state at saturating concentrations of maltose, with no detectable dynamics on the relevant timescale of < 10 ms.
In the lab of Don Lamb, we had previously worked on a highly dynamic component of the spliceosome, the U2 auxiliary factor 2 (U2AF2) [4]. U2AF2 is comprised of two folded domains that are connected by a flexible, disordered linker. As a result, the protein samples an ensemble of extended conformation. In contrast, when bound to RNA, U2AF2 assumes a stable structure as both domains interact with the RNA (see Figure 2 and below). The initial round of measurements for U2AF2 revealed several issues. Different to MalE, which we had shipped in ambient conditions, the lower stability of U2AF2 necessitated snap-freezing and shipment on dry ice. In some cases, either shipping delays caused a depletion of the dry ice supply or inadequate handling after thawing led to degradation of the samples, which we addressed by resending fresh samples. In addition, U2AF2 showed strong sticking to glass surfaces, which led to sample loss during the measurement in cases where the passivation guidelines were not followed.
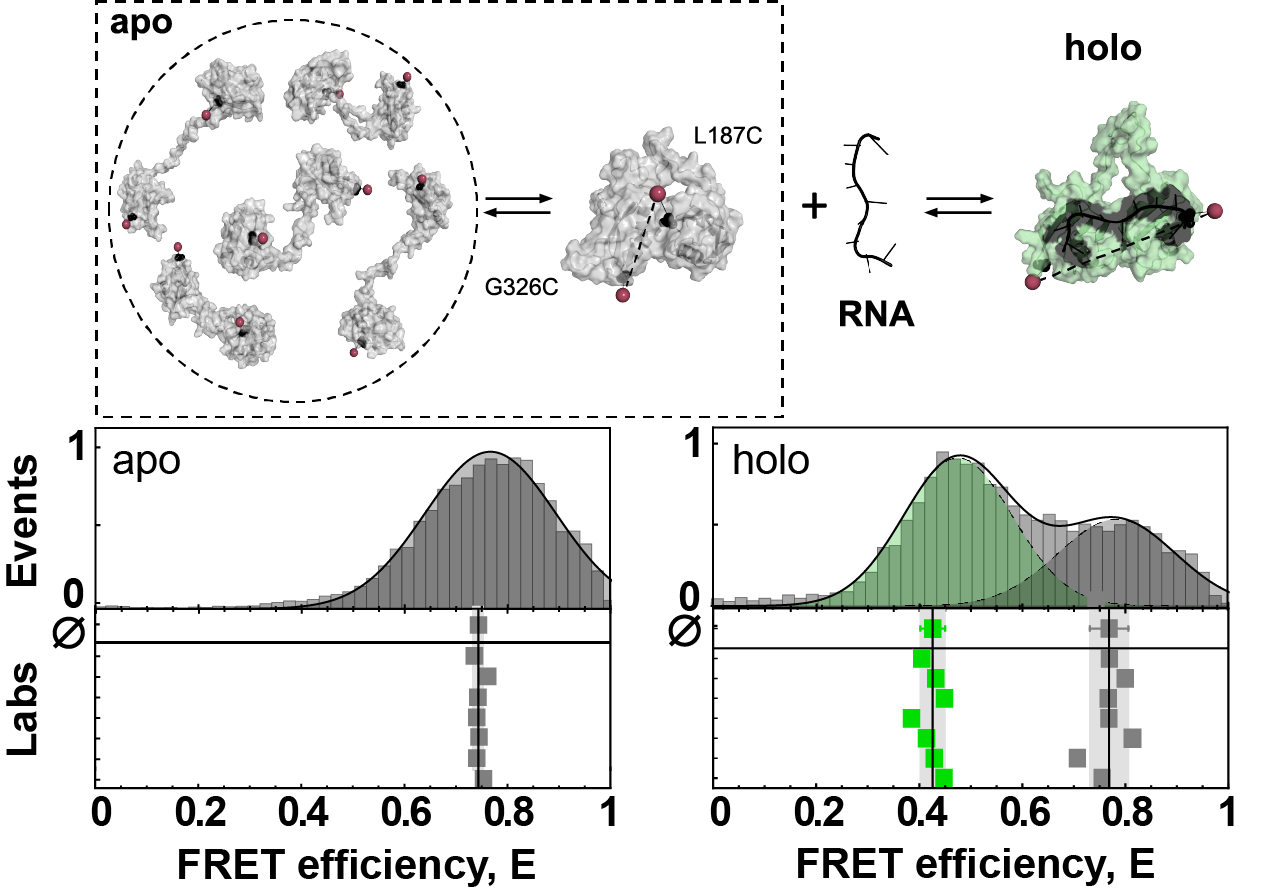
In the absence of RNA, U2AF2 samples an ensemble of detached conformation with occasional stable contacts between the domains. The RNA-bound state is static with a defined arrangement of the domains. The results of the smFRET experiments remain highly reproducible for the dynamic system.
Having solved these issues, we obtained good agreement of the reported FRET efficiency histograms for the dynamic U2AF2. However, we still observed larger variations compared to MalE, especially in the RNA-bound state (see Figure 2c), likely caused by temperature differences in the participating labs (affecting the dynamic equilibrium) or degradation of the RNA ligand. The highly dynamic RNA-free state seemed like an ideal system to test whether the high reproducibility of FRET efficiency histograms would also translate to the detection and quantification of conformational dynamics. Indeed, using two commonly applied methods for detection of dynamics we could show an excellent agreement even in terms of the timescales of the structural fluctuations (see Figure 4 and Figure 5e). Finally, there is more to be said about the complex dynamics of U2AF2 and how smFRET experiments can help understand them with state-of-the-art analysis methods. In the end, this analysis was condensed into Figure 6, but let it be noted that it took a large effort of first and corresponding authors alike to understand the U2AF2 system in detail.
All good then? Well, almost. Unexpectedly, we found that all labs consistently detected sub-millisecond dynamics for a particular mutant of MalE. After testing two additional mutants of MalE which did not show these dynamics, it became clear that the apparent dynamics must be an artifact of the dyes. Indeed, switching the dye pair diminished the false-positive dynamics for the problematic mutant (see Figure 5f). Ultimately, we found that the reason for this behavior were prominent sticking interactions of certain dyes that led to fluctuations of the apparent FRET efficiency. By quantifying the fluorescence anisotropy, this further allowed us to formulate an exclusion rule for filtering out ‘bad’ dyes and, ultimately, clean up our data to the point where we could detect small-scale distance fluctuations on the order of 5 Å for MalE. Additionally, by correcting for dye sticking in the distance estimation we could further improve our precision (Figure 5 c-d).
There is one more lesson to be learned from this study. We identified that the precision of FRET measurements is ultimately limited by the calibration, with the (infamous) correction factor γ having the largest influence (Figure 3e). Establishing robust and reproducible correction procedures is thus expected to further improve the accuracy and reproducibility, as we showed by having the reported data reanalyzed by a single person. While there is thus still room for improvement, we hope that the present study will raise the interest for smFRET experiments in the wider scientific community as an integral tool in dynamic structural biology [5].
Around the time of the first FRET challenge, we published our open-source software package that Anders had worked on over the course of his PhD [6]. Having spent significant amounts of time implementing advanced analysis methods, the hope at that time was that these efforts could benefit others in the field by making the analysis routines easily accessible to the community. (As irony has it, at that time we failed to raise the interest of the editors of Nature Methods to consider our publication.) Four years later it seems that the efforts were not in vain, as 9 of the 19 participating labs in the present study have applied our software for the analysis of their data. We like to think that this might have also played a role for the excellent agreement of the results.
Follow the Topic
-
Nature Methods

This journal is a forum for the publication of novel methods and significant improvements to tried-and-tested basic research techniques in the life sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Methods development in Cryo-ET and in situ structural determination
Publishing Model: Hybrid
Deadline: Jul 28, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in