Scalable, FACS-Free Genome-Wide Phenotypic Screening Identifies a Candidate Cancer Immunotherapy Modulator
Published in Bioengineering & Biotechnology

CRISPR technology has revolutionized the world of functional genomics and now makes it possible to test all of our 20,000 genes to find the ones required for cellular viability in a given setting. However, looking for genes that control more subtle phenotypes, like modulations in protein expression or modification, is much more challenging. This is because lots of time and money have to be spent on sorting lots of cells using FACS technology – which sorts cells one at a time based on the binding of a fluorescent antibody. If you ask researchers who perform these screens and watch dancing dots in the FACS control software for several consecutive days, only interrupted by changing buffer tanks or resolving clogs, they would likely use the word “painful”.
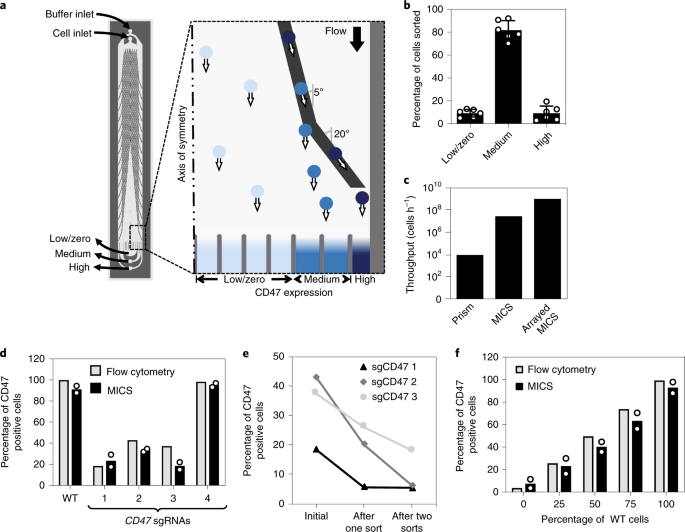
In our multidisciplinary research environment in Toronto, we started brainstorming about creating devices to ease the processing of such screens. The Moffat and Kelley labs quickly honed in on a microfluidic cell sorting approach to increase throughput. While the general idea seemed fairly straightforward, Peter Aldridge, the microfluidics expert, and I, the CRISPR postdoc, realized that the devil was in the details.
How many cells would we need to sort, and how many could we sort? Into how many expression bins? Which target protein should we look at? What cell line? For each of these questions, we designed, we optimized, we tested – and went back to the drawing board until we could check all the boxes. HAP1 cells are our workhorse, easy choice. CD47 is a candidate cancer immunotherapy target, that’s interesting. There are good antibodies allowing us to pick out cells with up- or down regulated CD47. Processing a billion cells per hour was the goal, and we finally got there as well.
The initial euphoria after our screen correctly identified all controls turned into us scratching our heads when staring at the hit list. Our top candidates seemed somewhat non-obvious and included an enzyme that cyclizes glutamine residues, QPCTL. How would this make any sense? So we did what we do best: another screen, this time the conventional way using FACS. But then it was obvious: QPCTL was a real hit, coming out at the top again.
From there, things started moving quickly – the more we studied QPCTL, the more interesting it became. We realized that QPCTL cyclized the N-terminal amino acid of CD47, and that we had picked this up because our capture antibody only binds to modified CD47. On top, the modification is critical for CD47’s interaction with SIRPA, a protein displayed by macrophages that reads CD47 as a “don’t eat me” signal.
In addition to having discovered an interesting aspect of immunotherapy target regulation, we now have in hand a powerful approach that allows phenotypic CRISPR screens to be processed much faster. For the near future, we are working towards higher parallelization to perform sets of screens investigating families of targets in panels of cell lines. We are extremely excited to see how this technology can help us to uncover new biology.
Follow the Topic
-
Nature Biomedical Engineering

This journal aspires to become the most prominent publishing venue in biomedical engineering by bringing together the most important advances in the discipline, enhancing their visibility, and providing overviews of the state of the art in each field.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in