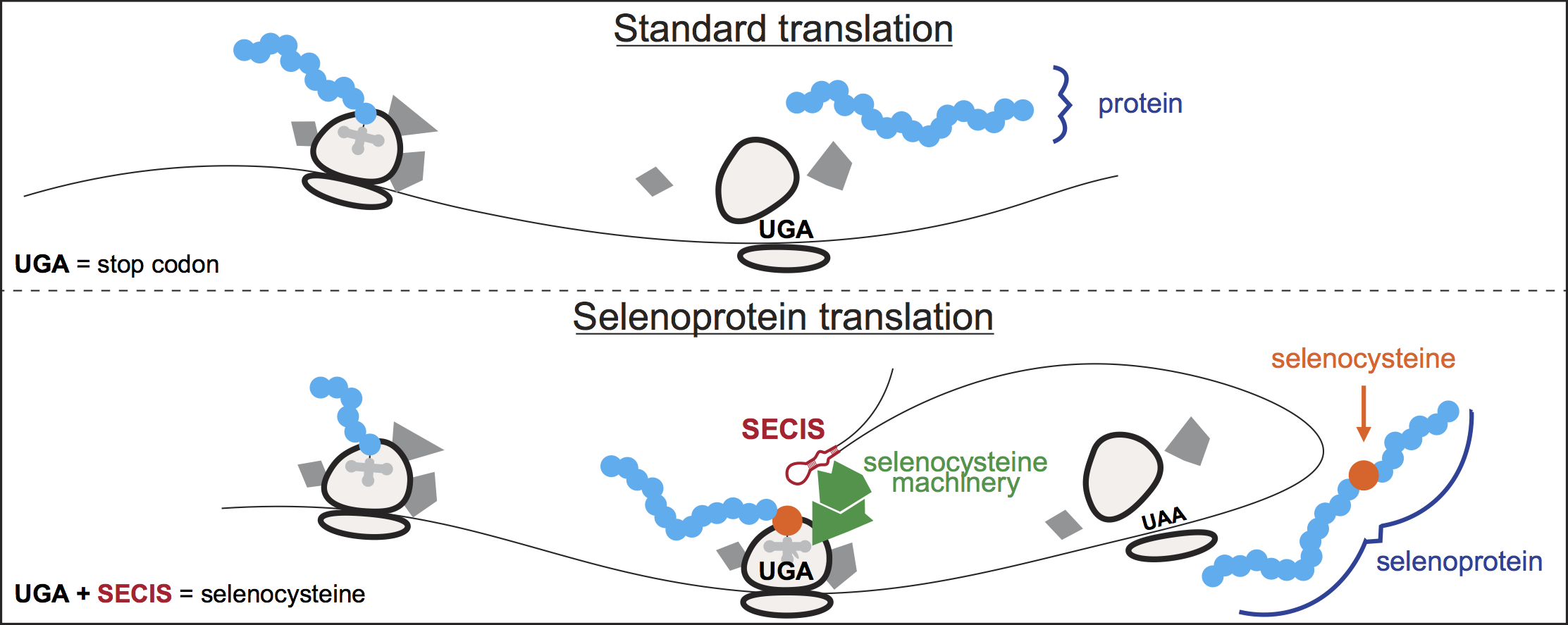
Translation is one of the most fundamental processes in biology, and it is ubiquitous to life. As we all learn in high-school, the ribosome reads each messenger RNA in triplets ('codons'), whose sequence determines which amino acids get incorporated in the synthesized protein. There are 64 possible triplets: 61 code for 20 different amino acids, while the remaining three act as stop signals during translation. Yet, even among biologists, few know that our own human cells utilize one extra amino acid: selenocysteine, dubbed the 21st amino acid. Selenocysteine is a special amino acid. Unlike standard amino acids, selenocysteine does not have a fully dedicated codon: instead, one of the stop codons, UGA, is recoded to selenocysteine. Translation is altered to insert selenocysteine for just a few 'selenoprotein' genes in the genome, marked by dedicated RNA structures (SECIS elements). This process is complex, inefficient and energetically expensive. Then, why do our cells take all this trouble?
While this remains an open question, biochemists point to the unique properties of the element selenium, which lays at the core of selenocysteine and enhances certain enzymatic activities. This question is particularly relevant because many species do not use selenocysteine at all. Typically, these species contain proteins that resemble our own selenoproteins but replace selenocysteine with cysteine, its most similar amino acid in the standard repertoire. The distribution of selenocysteine among living species reveals an insightful snapshot of evolution at work. Selenocysteine is well conserved in human and the rest of vertebrates; yet, various branches of the tree of life have dropped it along the way, and have thrived nevertheless. Humans have 25 selenoprotein genes; fruit flies has three; the worm Caenorhabditis elegans has only one. Land plants do not have any, but algae do. Among prokaryotes, selenoproteins are present in only ~20% of species. This scattered pattern testifies a dynamic evolutionary process acting on selenium utilization.
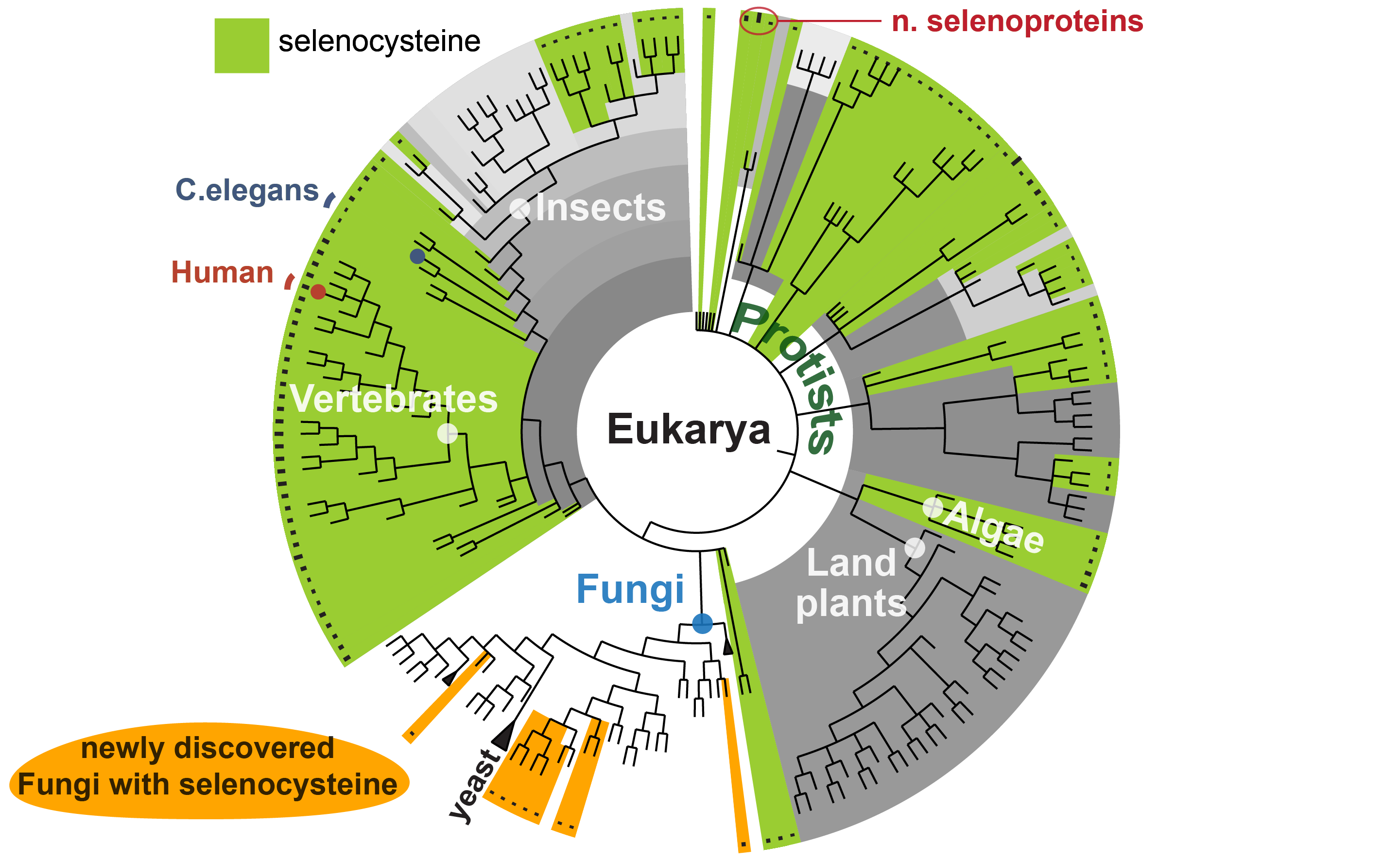
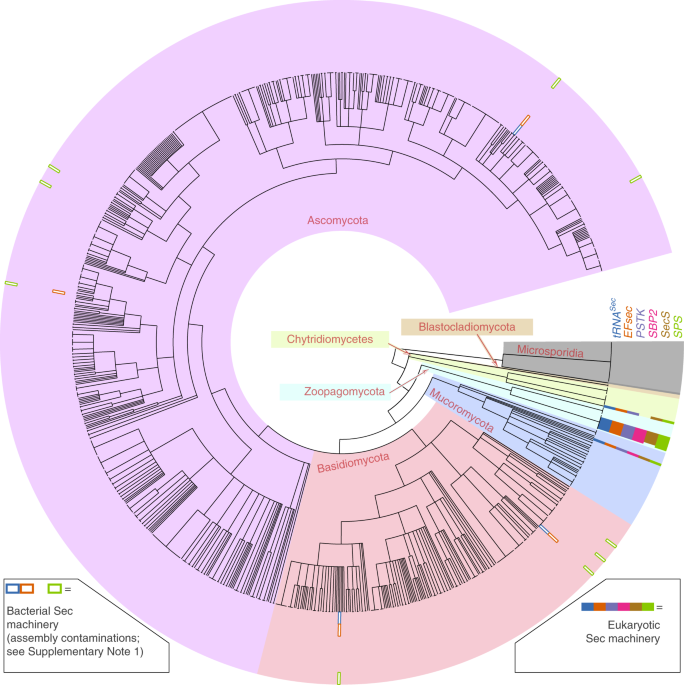
In our article, we shed new light on this process. Since yeast and other well studied fungi were found to lack selenoproteins, it was thought that selenocysteine was absent from the whole fungal kingdom. In fact, fungi were the only classically defined kingdom in biology that lacked selenoproteins entirely (since selenoprotein-containing organisms are known in Animalia, Plantae, Protista, Archaea, and Bacteria). Thanks to the rise of sequencing and the new outlook of genetic biodiversity at our disposal, we now know that this is not true: applying gene prediction and evolutionary analyses, we discovered selenocysteine machinery and selenoprotein genes in several newly sequenced species of fungi. These organisms belong to early-branching lineages, indicating that selenocysteine was originally present in fungi but it was then lost in the yeast branch. The fungal genes that we identified bear clear similarity to the previously identified selenoproteins of other species, including human. Yet, it appears that the mechanisms of selenocysteine recoding have diversified in fungi, manifested by frequent absence of canonical eukaryotic SECIS. Notably, the selenocysteine distribution across fungi implies that multiple independent events of selenoprotein losses occurred. The driving force of selenocysteine maintenance and loss in different lineages is still unclear, but our discovery in fungi brings us closer. Understanding why selenoproteins ceased to be important in fungi and other branches of the tree of life may tell us why, in contrast, it is so important for our own species, to define what makes selenium essential for human health.
Reference: Utilization of selenocysteine in early-branching fungal phyla. Marco Mariotti, Gustavo Salinas, Toni Gabaldón & Vadim N. Gladyshev. Nature Microbiology (2019). https://doi.org/10.1038/s41564-018-0354-9 Published: 11 February 2019.
Accessible at https://www.nature.com/articles/s41564-018-0354-9
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in