Sequencing-guided re-estimation and promotion of cultivability for environmental bacteria
Published in Microbiology, Protocols & Methods, and Genetics & Genomics
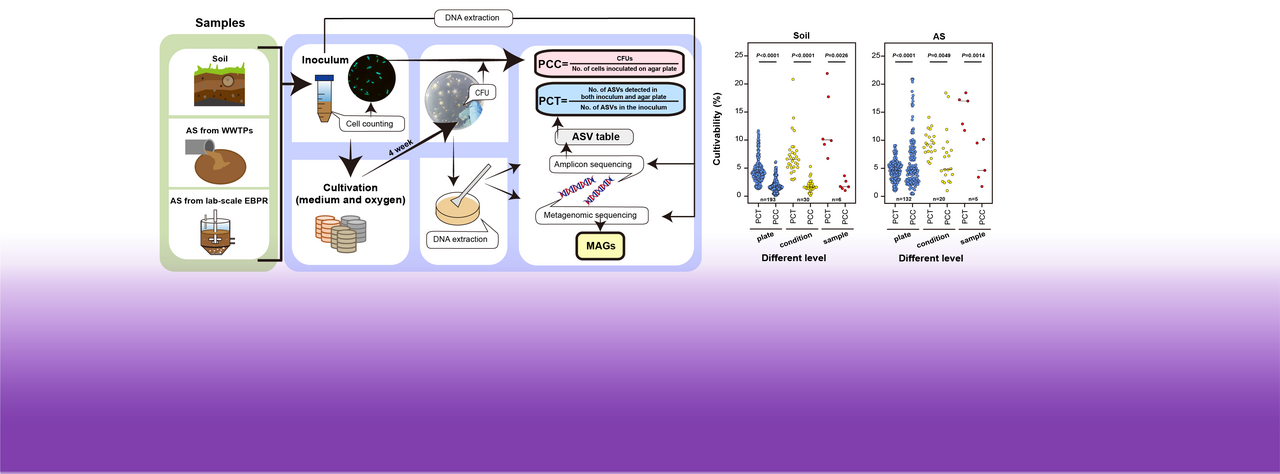
The mystery of the 'great plate count anomaly' in environmental microbes has persisted for over a century. This phenomenon refers to the observation that the number of inoculated cells counted microscopically is often 10-1000-fold higher than the number of colony-forming units (CFUs) observed on an agar plate, in a typical environmental sample. Therefore, the widely accepted paradigm that 'only 1% of microorganisms can be cultured'—meaning the vast majority cannot—has become commonplace. However, most previous estimates of bacterial cultivability have primarily focused on the proportion of cultivable cells (PCCs).
However, from the perspective of microbial resources and diversity, more attention should be given to the proportion of cultivable taxa (PCTs) among all taxa. In 2019, Martiny conducted a PCT estimation based on a 16S rRNA gene database, raising concerns about the potential underestimation of cultivability at the taxonomic level. However, a subsequent discussion argued that this conclusion was based on an inherently biased concept, emphasizing that it is impossible to determine a microbe's cultivability until it has been successfully cultivated under specific conditions. Due to the significant mismatch between laboratory culture conditions and the natural growth conditions of microorganisms, most microorganisms remain 'unculturable' or 'uncultured.' This includes the so-called 'microbial dark matter,' which encompasses microorganisms that cannot be cultured and represent strains at higher taxonomic levels, such as phyla or class.
Our research re-evaluated the cultivability of environmental bacteria, comparing PCT and PCC, revealing that cultivability has been generally underestimated in the past. Additionally, this study proposes treating cultivable traits under certain conditions as phenotypes and using sequencing data to explore information on cultivable bacterial groups and their genomic metabolic potential, which can guide the isolation and cultivation of specific microorganisms.

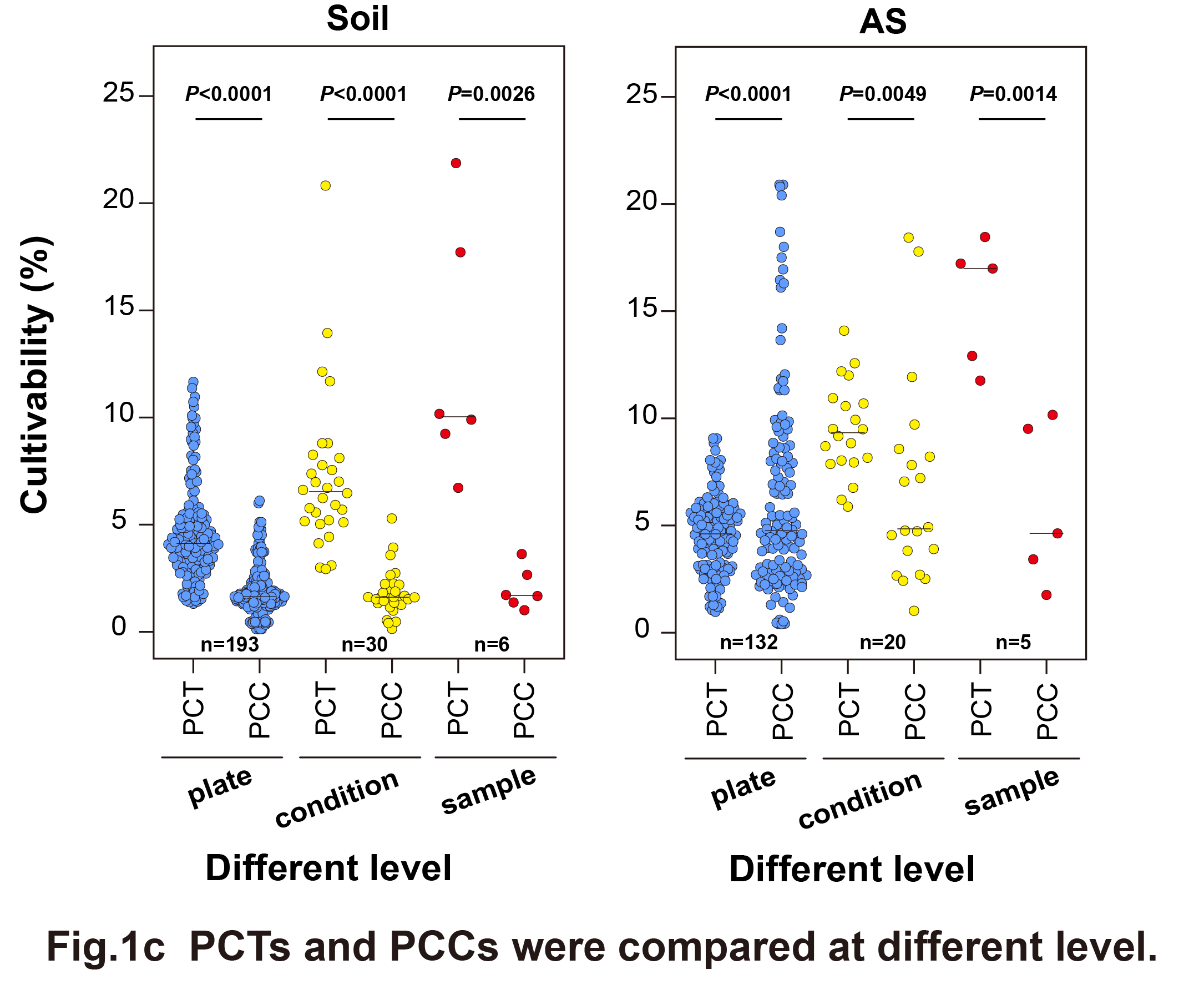
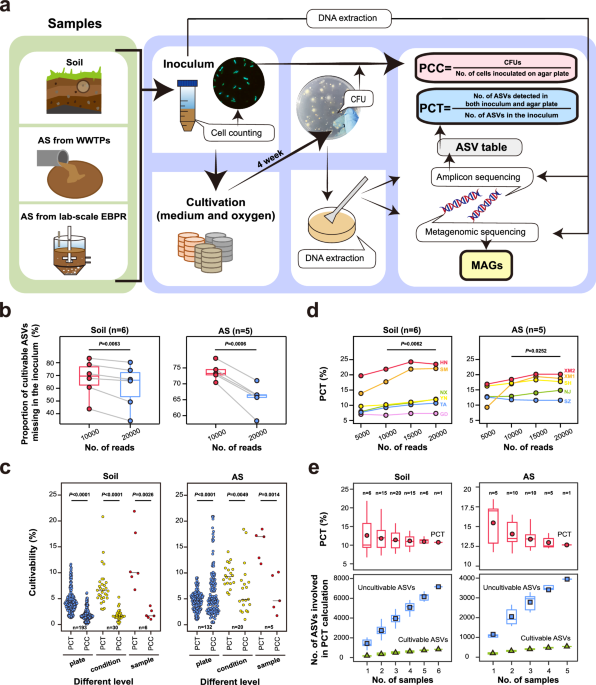
Simplified culturomics methods, involving different oxygen conditions and mediums, were applied to bacteria from two typical environmental samples (6 soil samples and 5 activated sludge samples). Cell-level cultivability was assessed using traditional microscopic counts and CFU counts. Sequencing provided bacterial group data from both the original samples and plate cultures, allowing the calculation of the proportion of cultivable taxa (PCT) (Fig. 1a). The results showed that PCT was significantly higher than PCC (approximately 2.8 to 6 times) under full culture conditions (Fig. 1c), indicating that cell-level measurements underestimate the cultivability diversity of environmental bacteria.
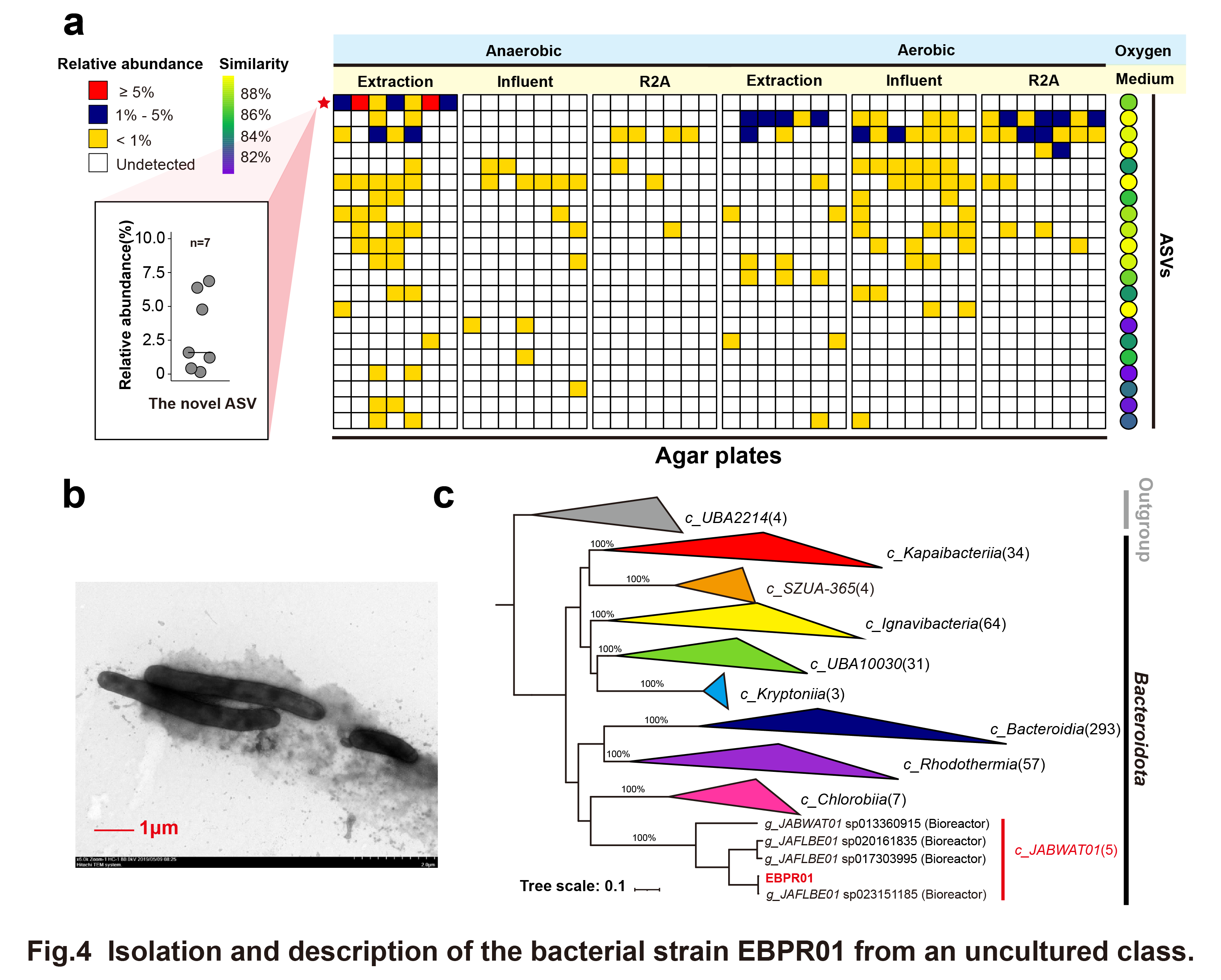
The 'cultivable/uncultivable' features under specific culture conditions are considered bacterial phenotypes. This phenotypic information can enhance the efficiency of isolating and culturing key microorganisms in two ways: 1) By using taxonomic information of cultivable bacteria, it can identify which samples and culture conditions favor the growth of specific bacterial groups of interest, helping to target the desired bacteria; 2) By comparing the genomic information of uncultivable and cultivable bacteria within a limited phylogenetic range (focused on one bacterial order in this study), key differential genes can be identified, allowing researchers to infer potential improvements to culture conditions. In this study, a new class-representative strain, EBPR01, was successfully isolated from an activated sludge sample sourced from a reactor using special culture conditions (extraction medium and anaerobic condition) (Fig. 4).
Also, we found that vitamin B12 synthesis genes were generally absent in Burkholderiales, and supplementing the medium with vitamin B12 significantly increased the cultivable diversity of this group (Fig. 6b, c).

In conclusion, this study proposed a novel strategy for evaluating and enhancing the cultivability of environmental bacteria using sequencing-based methods. The innovative combination of sequencing technology with culture techniques may help uncover more key factors that enlighten the microbial dark matter to 'forsake darkness for light,' narrowing the gap between natural microbial diversity and what can be cultured in the lab. Expanding our ability to culture a wider range of microbes opens up new possibilities for discoveries in fields such as bioremediation, biotechnology, and medicine.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in