Simplicity is the ultimate sophistication: self-aggregation in the heart
Published in Protocols & Methods

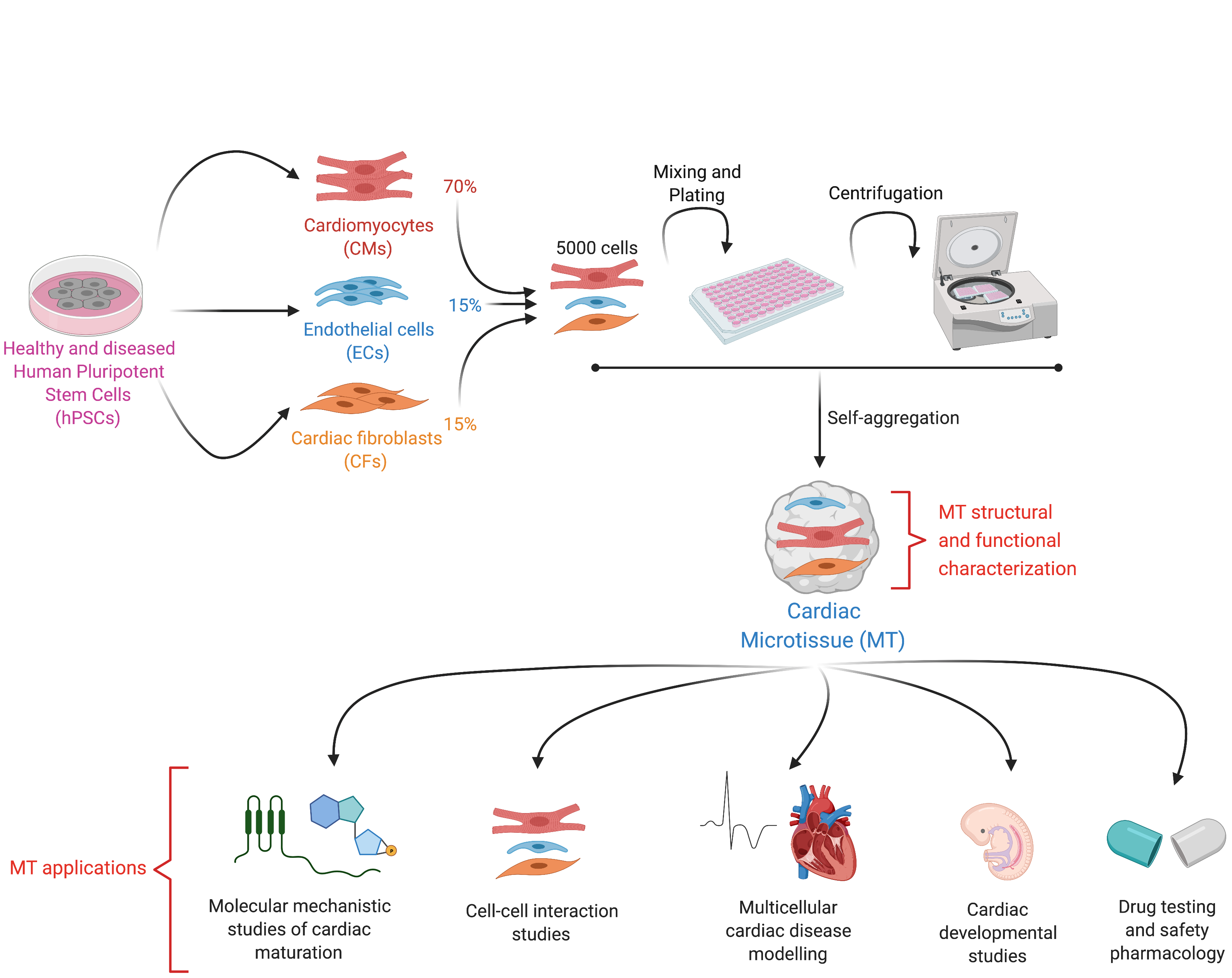
In the last decade, scientists discovered how to grow human pluripotent stem cells (hPSCs) in aggregates of thousands and millions of cells that, after culture with specific growth factors or small molecules, look and act increasingly like organs of the human body. These so-called "organoids" are currently used to model human diseases, screen- and pharmacologically profile- drugs, and to increase our understanding of human development and even human evolution1.
By definition, an organoid is a self-organizing aggregate of cells that 1) contains multiple organ-specific cell types; 2) is capable of recapitulating some specific functions of the organ; and 3) is spatially organized much like an organ2. In the Mummery Lab, we applied these principles to develop an organoid-like model of the human heart not through self-organization but by assembling pre-differentiated cell types. These human heart models contain the three major cell types present in the organ: cardiomyocytes (CMs), the muscle cells that contract; cardiac endothelial cells (ECs), the cells that supply oxygen and nutrients and cardiac fibroblasts (CFs), the cells of the connective tissue that primarily produce the extracellular matrix. However, because self-organization in our model does not occur from the hPSC stage (a key feature of true organoids), but instead, fully differentiated cells are mixed and form a 3-D tissue ~500 micrometer in diameter, we decided to call our model "cardiac microtissue".
This journey began at the start of my PhD, in 2015, in the place of the lab that I enjoy the most, the cell culture room. One Friday afternoon, one of my supervisors (Milena Bellin) and I combined some CMs and ECs (derived from hPSCs) in a 96 (V) bottom plate – we had so many in the lab as nobody was using them anymore. We left the plate in the incubator over the weekend and quickly checked on Monday morning, right before our regular 9am lab meeting. Surprise! The CMs and ECs had aggregated beautifully into the shape of a sphere that was spontaneously beating. Using a human embryonic stem cell (hESC)-eGFP reporter line3, we could visualize the CM population in green and record videos of the beating spheres (Figure 1). What we tried on that day was nothing sophisticated or extremely challenging. It was just a combination of the correct cell numbers, media composition and plate format, which led to the beginning of a beautiful scientific adventure. A great Monday to start the week! The lesson I learnt from that: the simplest things in scientific life may be the ones that work the best.

Figure 1: Where we started - first microtissue model containing CMs and ECs generated from a (hESC)-eGFP reporter line3
We published the first version of our microtissue model in 2016 as part of the Organoids Special Issue in Development4, and we later described the step-by-step protocol for this5. Through this work, we found that ECs, the area of Valeria Orlova in the lab, were essential to induce some aspects of CM maturity. This lack of maturity was a crucial limitation of hPSC-derivatives at that time, as it meant that they resemble fetal cells rather than mature, adult-like CMs. However, the CM maturity induced by the 3-D environment and the presence of ECs was still incomplete. It was only by differentiating and incorporating CFs into the microtissue that we could generate CMs with electrophysiological, structural, mechanical, and metabolic maturity6. This level of maturity had rarely been reported before without extended electrical pacing in culture, and our method had the added benefit of being small-scale and high throughput for downstream applications6. CFs are critical for normal cardiac function and remodelling during pathological conditions such as Arrhythmogenic Cardiomyopathy (ACM). This cardiac disorder is characterized by arrhythmia and replacement of the myocardium with fibro-fatty tissue. While CFs are the largest cell population of the heart, little is known about their role in cardiac maturation and cardiac disorders such as ACM. At this point, Giulia Campostrini, Viviana Meraviglia and I joined efforts to understand how CFs contribute to the maturation of microtissues functionally (Giulia) and how they contribute to the ACM disease phenotype (Viviana). Using our three-cell type microtissue, we found that the dialogue among all three cell types was essential for the highest degree of maturation in the CMs6. CFs induced maturation by expressing connexin 43 (CX43) gap junctions and cAMP-pathway activation6. Maturity was also retained after single-cell dissociation of the microtissue, allowing further functional characterization of the model6. Our findings also provided evidence of non-CM contribution to ACM pathogenesis, thus demonstrating the utility of our microtissue system for modeling diseases not autonomous to CMs6.
We are now extremely excited to share our new Nature Protocol with a broad group of end-users.
Our protocol:
- Supports CM maturity and allows replacement of one or more of the three cell types in the microtissue with isogenic variants bearing disease mutations
- Is a simple and versatile platform for modelling multicellular cardiac diseases, understanding multicellular heart conditions, identifying therapeutic targets and predicting drug efficacy, as recently published7.
- Is scaffold-free, easy to fabricate, low cost, and does not require specific tissue engineering expertise, specialized apparatus or analysis tools. The cardiac cells can be frozen "ready to go" for experiments. Hence, it can be helpful for a wider group of researchers.
The current protocol is used by technicians, PhD students and postdoctoral fellows in the Mummery Lab, with several follow up manuscripts using the method currently being prepared and/or submitted for peer review. Our goal in writing this protocol is to provide researchers with a robust and reproducible platform to implement in their research laboratory. Due to the broad applicability, we hope our microtissue platform will be readily adopted by other research laboratories, thus accelerating cardiovascular research.
References
1. Trujillo, C. A. et al. Reintroduction of the archaic variant of NOVA1 in cortical organoids alters neurodevelopment. Science 371, doi:10.1126/science.aax2537 (2021).
2. Lancaster, M. A. & Knoblich, J. A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125, doi:10.1126/science.1247125 (2014).
3. Elliott, D. A. et al. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods 8, 1037-1040, doi:10.1038/nmeth.1740 (2011).
4. Giacomelli, E. et al. Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development 144, 1008-1017, doi:10.1242/dev.143438 (2017).
5. Giacomelli, E., Bellin, M., Orlova, V. V. & Mummery, C. L. Co-Differentiation of Human Pluripotent Stem Cells-Derived Cardiomyocytes and Endothelial Cells from Cardiac Mesoderm Provides a Three-Dimensional Model of Cardiac Microtissue. Curr Protoc Hum Genet 95, 21 29 21-21 29 22, doi:10.1002/cphg.46 (2017).
6. Giacomelli, E. et al. Human-iPSC-Derived Cardiac Stromal Cells Enhance Maturation in 3D Cardiac Microtissues and Reveal Non-cardiomyocyte Contributions to Heart Disease. Cell Stem Cell 26, 862-879 e811, doi:10.1016/j.stem.2020.05.004 (2020).
7. Qiao, X. et al. Uncoupling DNA damage from chromatin damage to detoxify doxorubicin. Proc Natl Acad Sci U S A 117, 15182-15192, doi:10.1073/pnas.1922072117 (2020).
Acknowledgment: BioRender (https://BioRender.com)
Follow the Topic
-
Nature Protocols

This journal publishes secondary research articles and covers new techniques and technologies, as well as established methods, used in all fields of the biological, chemical and clinical sciences.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in