Size matters: supersized bacterial secretion system
Published in Microbiology

One of the many ways bacteria can kill their competitors is using the Type VI secretion system (T6SS). This nanomachine contracts its spring-like component called sheath to push a long tube with associated toxins into various target cells. The sheath assembles across the whole bacterial cell and can be visualized by live-cell fluorescence microscopy. Our lab is interested in understanding the mode of action of the T6SS in many different bacteria. For example, we want to know where and how are the sheath subunits added to the assembling T6SS. To address this question experimentally, we came up with a strategy to use a strong laser beam to photobleach a section of an assembling sheath in live cells and then observe the localization of the bleached section during continuing sheath assembly. Different mechanisms of sheath assembly would be possible to distinguish by subsequent image analysis. However, there was one problem. Even though a T6SS sheath usually assembles across a whole cell, laser beam is impossible to focus to a spot smaller than a diameter of an average bacterial cell thus preventing partial photobleaching of the sheath. Therefore, our plan was put on hold as it became clear that we needed a different experimental approach. Or … as it turned out later … we just needed bigger cells!
To study T6SS function, we often use plasmids encoding resistance to ampicillin to express different T6SS components. To ensure that the plasmids are present in all bacteria, we grow them in the presence of ampicillin. The idea is that the cells that lose the plasmid will become sensitive to the antibiotic and lyse because their cell wall synthesis would be inhibited. As many times before, we were watching Vibrio cholerae cells assembling its T6SS and between the normal rod-shaped Vibrio cells, we noticed few round cells, which likely recently lost the plasmid and thus lacked their cell wall. At first, we ignored those round cells but after collecting several time-lapse movies with the goal to look at the dynamics of the T6SS, we made a curious observation that the round cells were alive and assembled T6SS similarly to their normal looking neighbors. That was truly unexpected because bacteria usually die and lyse without their cell wall.
We became curious how large the Vibrio cells may become before lysing. With no goal in mind, just to have some fun, we decided to watch a non-resistant Vibrio strain grow and assemble its T6SS in the presence of ampicillin. We were astonished to see that most of the bacteria lost their shape after few tens of minutes, stopped dividing and then kept growing bigger and bigger over time. Most importantly, the T6SS sheath was still dynamic and assembled across the round cells, occasionally reaching lengths over ten micrometers.
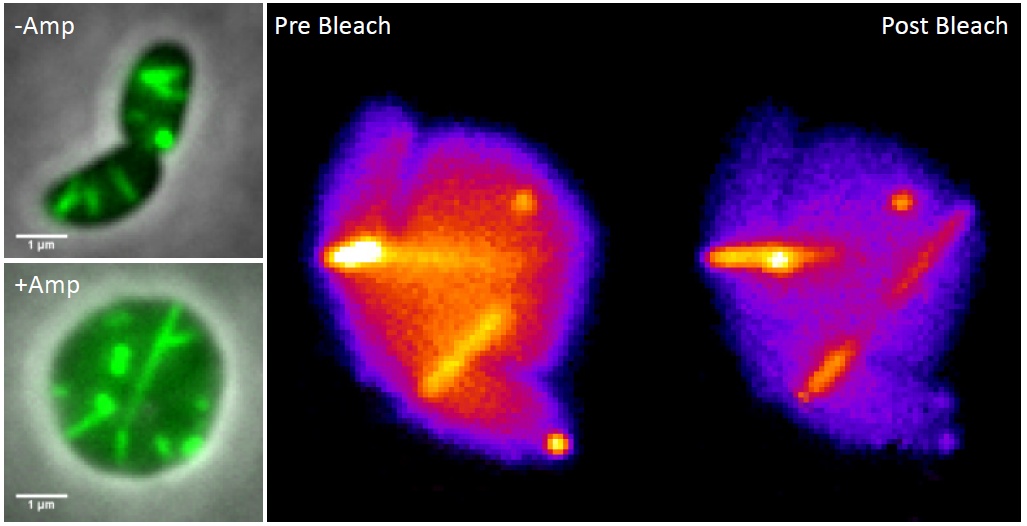
Left: T6SS sheath (green) visualized in rod-shaped and ampicillin treated V. cholerae cells.
Right: Partial photobleaching of an assembling T6SS sheath in V. cholerae spheroplast.
Watching long T6SS sheaths was a lot of fun and we realized that the large cells could allow us to come back to our question about sheath assembly. Indeed, Andrea, Julius and Lin elegantly showed that large round cells lacking their cell wall assemble functional T6SS and that the soluble sheath subunits are added to the growing polymer at the end distal from the T6SS membrane attachment. Our study shows that it pays off to keep eyes open and think about how to use new curious observations to revisit old questions.
Our Nature Communications paper is here: http://go.nature.com/2tbvjLC
Make sure you look at the supplementary videos!





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in